Kristin Schmidt
A Large Encoder-Decoder Family of Foundation Models For Chemical Language
Jul 24, 2024Abstract:Large-scale pre-training methodologies for chemical language models represent a breakthrough in cheminformatics. These methods excel in tasks such as property prediction and molecule generation by learning contextualized representations of input tokens through self-supervised learning on large unlabeled corpora. Typically, this involves pre-training on unlabeled data followed by fine-tuning on specific tasks, reducing dependence on annotated datasets and broadening chemical language representation understanding. This paper introduces a large encoder-decoder chemical foundation models pre-trained on a curated dataset of 91 million SMILES samples sourced from PubChem, which is equivalent to 4 billion of molecular tokens. The proposed foundation model supports different complex tasks, including quantum property prediction, and offer flexibility with two main variants (289M and $8\times289M$). Our experiments across multiple benchmark datasets validate the capacity of the proposed model in providing state-of-the-art results for different tasks. We also provide a preliminary assessment of the compositionality of the embedding space as a prerequisite for the reasoning tasks. We demonstrate that the produced latent space is separable compared to the state-of-the-art with few-shot learning capabilities.
Beyond Chemical Language: A Multimodal Approach to Enhance Molecular Property Prediction
Jun 22, 2023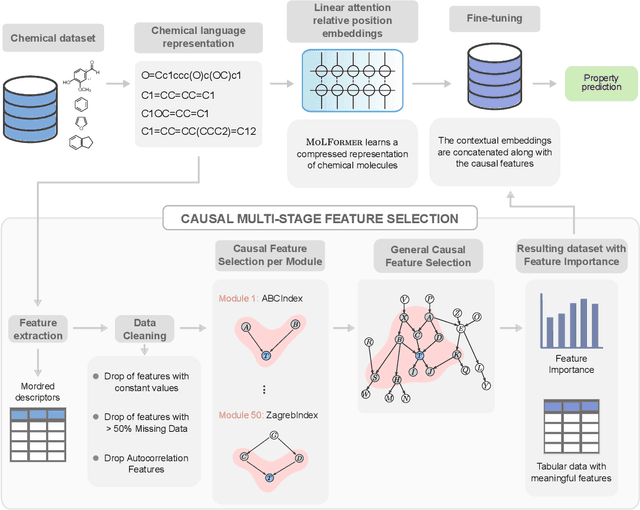
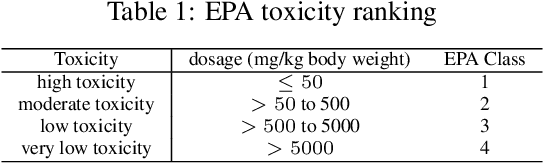
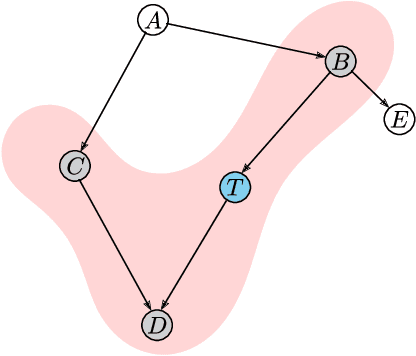
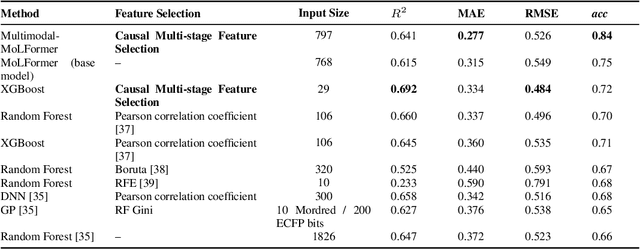
Abstract:We present a novel multimodal language model approach for predicting molecular properties by combining chemical language representation with physicochemical features. Our approach, MULTIMODAL-MOLFORMER, utilizes a causal multistage feature selection method that identifies physicochemical features based on their direct causal effect on a specific target property. These causal features are then integrated with the vector space generated by molecular embeddings from MOLFORMER. In particular, we employ Mordred descriptors as physicochemical features and identify the Markov blanket of the target property, which theoretically contains the most relevant features for accurate prediction. Our results demonstrate a superior performance of our proposed approach compared to existing state-of-the-art algorithms, including the chemical language-based MOLFORMER and graph neural networks, in predicting complex tasks such as biodegradability and PFAS toxicity estimation. Moreover, we demonstrate the effectiveness of our feature selection method in reducing the dimensionality of the Mordred feature space while maintaining or improving the model's performance. Our approach opens up promising avenues for future research in molecular property prediction by harnessing the synergistic potential of both chemical language and physicochemical features, leading to enhanced performance and advancements in the field.
Toward Human-AI Co-creation to Accelerate Material Discovery
Nov 05, 2022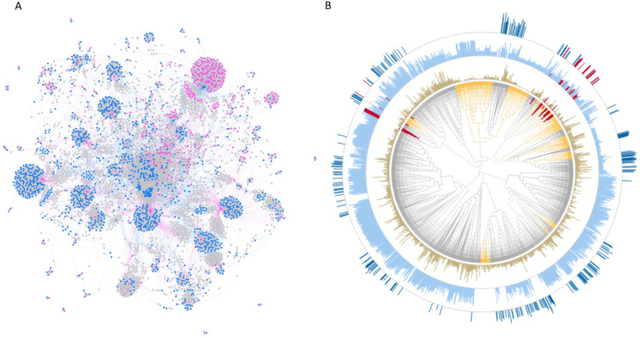
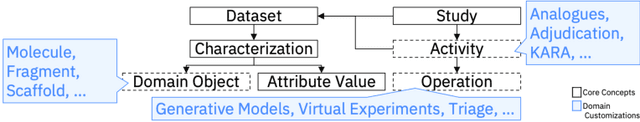
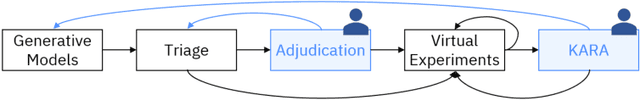
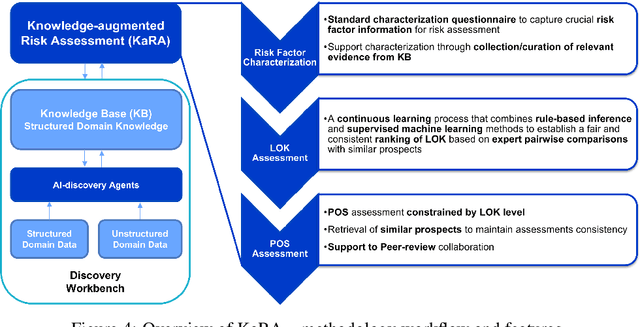
Abstract:There is an increasing need in our society to achieve faster advances in Science to tackle urgent problems, such as climate changes, environmental hazards, sustainable energy systems, pandemics, among others. In certain domains like chemistry, scientific discovery carries the extra burden of assessing risks of the proposed novel solutions before moving to the experimental stage. Despite several recent advances in Machine Learning and AI to address some of these challenges, there is still a gap in technologies to support end-to-end discovery applications, integrating the myriad of available technologies into a coherent, orchestrated, yet flexible discovery process. Such applications need to handle complex knowledge management at scale, enabling knowledge consumption and production in a timely and efficient way for subject matter experts (SMEs). Furthermore, the discovery of novel functional materials strongly relies on the development of exploration strategies in the chemical space. For instance, generative models have gained attention within the scientific community due to their ability to generate enormous volumes of novel molecules across material domains. These models exhibit extreme creativity that often translates in low viability of the generated candidates. In this work, we propose a workbench framework that aims at enabling the human-AI co-creation to reduce the time until the first discovery and the opportunity costs involved. This framework relies on a knowledge base with domain and process knowledge, and user-interaction components to acquire knowledge and advise the SMEs. Currently,the framework supports four main activities: generative modeling, dataset triage, molecule adjudication, and risk assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge