Kristen Morse
Masked Autoencoders for Microscopy are Scalable Learners of Cellular Biology
Apr 16, 2024



Abstract:Featurizing microscopy images for use in biological research remains a significant challenge, especially for large-scale experiments spanning millions of images. This work explores the scaling properties of weakly supervised classifiers and self-supervised masked autoencoders (MAEs) when training with increasingly larger model backbones and microscopy datasets. Our results show that ViT-based MAEs outperform weakly supervised classifiers on a variety of tasks, achieving as much as a 11.5% relative improvement when recalling known biological relationships curated from public databases. Additionally, we develop a new channel-agnostic MAE architecture (CA-MAE) that allows for inputting images of different numbers and orders of channels at inference time. We demonstrate that CA-MAEs effectively generalize by inferring and evaluating on a microscopy image dataset (JUMP-CP) generated under different experimental conditions with a different channel structure than our pretraining data (RPI-93M). Our findings motivate continued research into scaling self-supervised learning on microscopy data in order to create powerful foundation models of cellular biology that have the potential to catalyze advancements in drug discovery and beyond.
Masked autoencoders are scalable learners of cellular morphology
Sep 27, 2023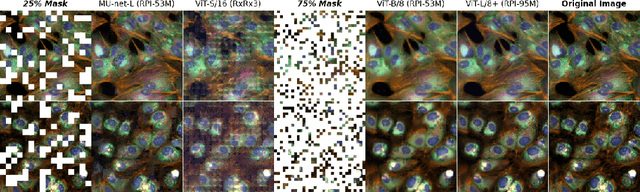
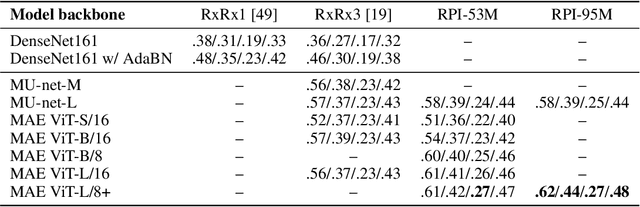
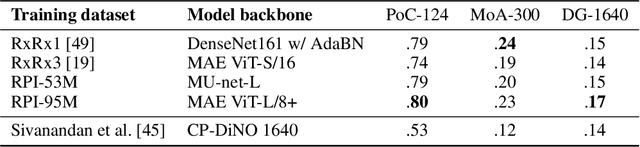
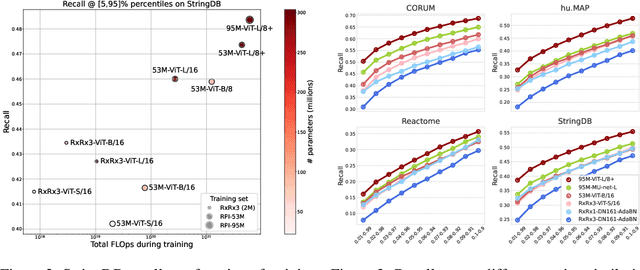
Abstract:Inferring biological relationships from cellular phenotypes in high-content microscopy screens provides significant opportunity and challenge in biological research. Prior results have shown that deep vision models can capture biological signal better than hand-crafted features. This work explores how weakly supervised and self-supervised deep learning approaches scale when training larger models on larger datasets. Our results show that both CNN- and ViT-based masked autoencoders significantly outperform weakly supervised models. At the high-end of our scale, a ViT-L/8 trained on over 3.5-billion unique crops sampled from 95-million microscopy images achieves relative improvements as high as 28% over our best weakly supervised models at inferring known biological relationships curated from public databases.
Learning State-Dependent Losses for Inverse Dynamics Learning
Mar 12, 2020



Abstract:Being able to quickly adapt to changes in dynamics is paramount in model-based control for object manipulation tasks. In order to influence fast adaptation of the inverse dynamics model's parameters, data efficiency is crucial. Given observed data, a key element to how an optimizer updates model parameters is the loss function. In this work, we propose to apply meta-learning to learn structured, state-dependent loss functions during a meta-training phase. We then replace standard losses with our learned losses during online adaptation tasks. We evaluate our proposed approach on inverse dynamics learning tasks, both in simulation and on real hardware data. In both settings, the structured learned losses improve online adaptation speed, when compared to standard, state-independent loss functions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge