Konstantina S. Nikita
Comparative assessment of fairness definitions and bias mitigation strategies in machine learning-based diagnosis of Alzheimer's disease from MR images
May 29, 2025Abstract:The present study performs a comprehensive fairness analysis of machine learning (ML) models for the diagnosis of Mild Cognitive Impairment (MCI) and Alzheimer's disease (AD) from MRI-derived neuroimaging features. Biases associated with age, race, and gender in a multi-cohort dataset, as well as the influence of proxy features encoding these sensitive attributes, are investigated. The reliability of various fairness definitions and metrics in the identification of such biases is also assessed. Based on the most appropriate fairness measures, a comparative analysis of widely used pre-processing, in-processing, and post-processing bias mitigation strategies is performed. Moreover, a novel composite measure is introduced to quantify the trade-off between fairness and performance by considering the F1-score and the equalized odds ratio, making it appropriate for medical diagnostic applications. The obtained results reveal the existence of biases related to age and race, while no significant gender bias is observed. The deployed mitigation strategies yield varying improvements in terms of fairness across the different sensitive attributes and studied subproblems. For race and gender, Reject Option Classification improves equalized odds by 46% and 57%, respectively, and achieves harmonic mean scores of 0.75 and 0.80 in the MCI versus AD subproblem, whereas for age, in the same subproblem, adversarial debiasing yields the highest equalized odds improvement of 40% with a harmonic mean score of 0.69. Insights are provided into how variations in AD neuropathology and risk factors, associated with demographic characteristics, influence model fairness.
Sustaining model performance for covid-19 detection from dynamic audio data: Development and evaluation of a comprehensive drift-adaptive framework
Sep 28, 2024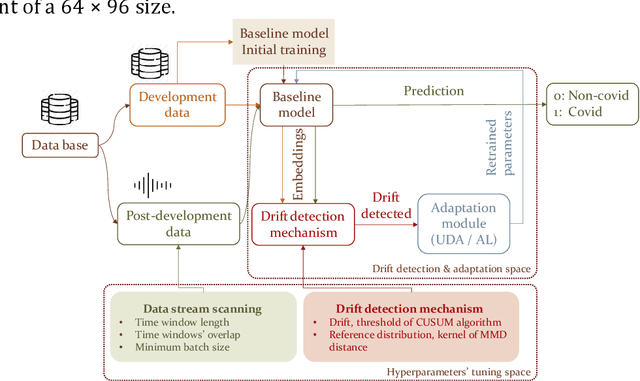
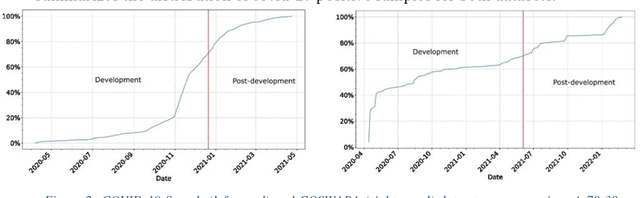
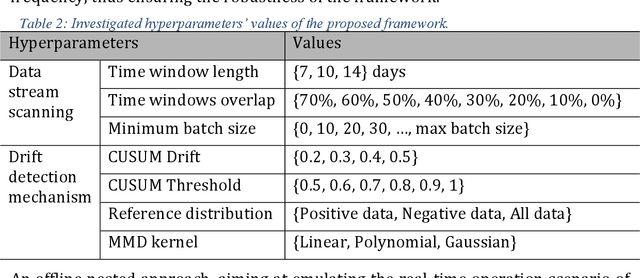
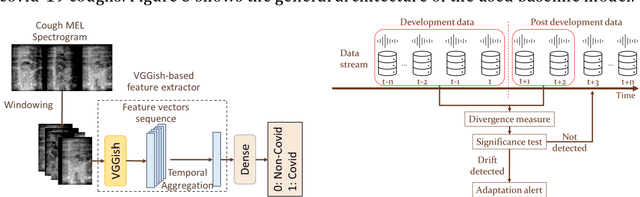
Abstract:Background: The COVID-19 pandemic has highlighted the need for robust diagnostic tools capable of detecting the disease from diverse and evolving data sources. Machine learning models, especially convolutional neural networks (CNNs), have shown promise. However, the dynamic nature of real-world data can lead to model drift, where performance degrades over time as the underlying data distribution changes. Addressing this challenge is crucial to maintaining accuracy and reliability in diagnostic applications. Objective: This study aims to develop a framework that monitors model drift and employs adaptation mechanisms to mitigate performance fluctuations in COVID-19 detection models trained on dynamic audio data. Methods: Two crowd-sourced COVID-19 audio datasets, COVID-19 Sounds and COSWARA, were used. Each was divided into development and post-development periods. A baseline CNN model was trained and evaluated using cough recordings from the development period. Maximum mean discrepancy (MMD) was used to detect changes in data distributions and model performance between periods. Upon detecting drift, retraining was triggered to update the baseline model. Two adaptation approaches were compared: unsupervised domain adaptation (UDA) and active learning (AL). Results: UDA improved balanced accuracy by up to 22% and 24% for the COVID-19 Sounds and COSWARA datasets, respectively. AL yielded even greater improvements, with increases of up to 30% and 60%, respectively. Conclusions: The proposed framework addresses model drift in COVID-19 detection, enabling continuous adaptation to evolving data. This approach ensures sustained model performance, contributing to robust diagnostic tools for COVID-19 and potentially other infectious diseases.
The smarty4covid dataset and knowledge base: a framework enabling interpretable analysis of audio signals
Jul 11, 2023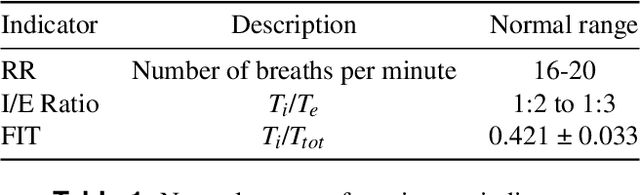
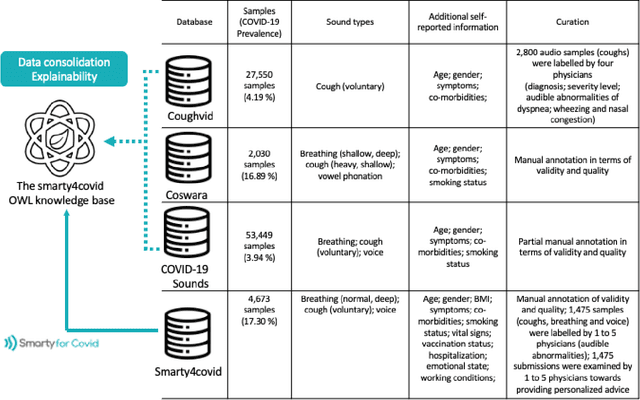

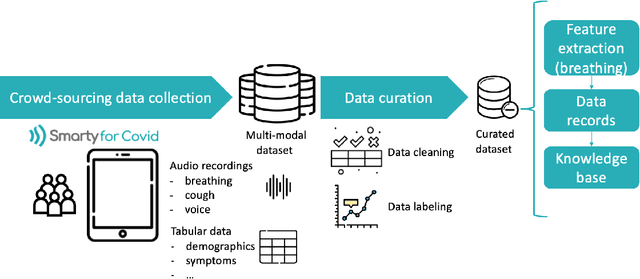
Abstract:Harnessing the power of Artificial Intelligence (AI) and m-health towards detecting new bio-markers indicative of the onset and progress of respiratory abnormalities/conditions has greatly attracted the scientific and research interest especially during COVID-19 pandemic. The smarty4covid dataset contains audio signals of cough (4,676), regular breathing (4,665), deep breathing (4,695) and voice (4,291) as recorded by means of mobile devices following a crowd-sourcing approach. Other self reported information is also included (e.g. COVID-19 virus tests), thus providing a comprehensive dataset for the development of COVID-19 risk detection models. The smarty4covid dataset is released in the form of a web-ontology language (OWL) knowledge base enabling data consolidation from other relevant datasets, complex queries and reasoning. It has been utilized towards the development of models able to: (i) extract clinically informative respiratory indicators from regular breathing records, and (ii) identify cough, breath and voice segments in crowd-sourced audio recordings. A new framework utilizing the smarty4covid OWL knowledge base towards generating counterfactual explanations in opaque AI-based COVID-19 risk detection models is proposed and validated.
Interpretability methods of machine learning algorithms with applications in breast cancer diagnosis
Feb 04, 2022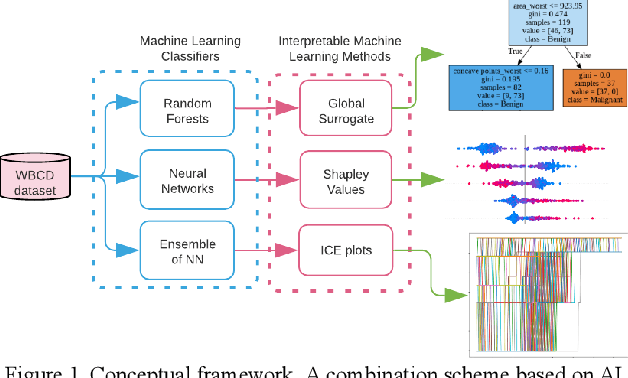
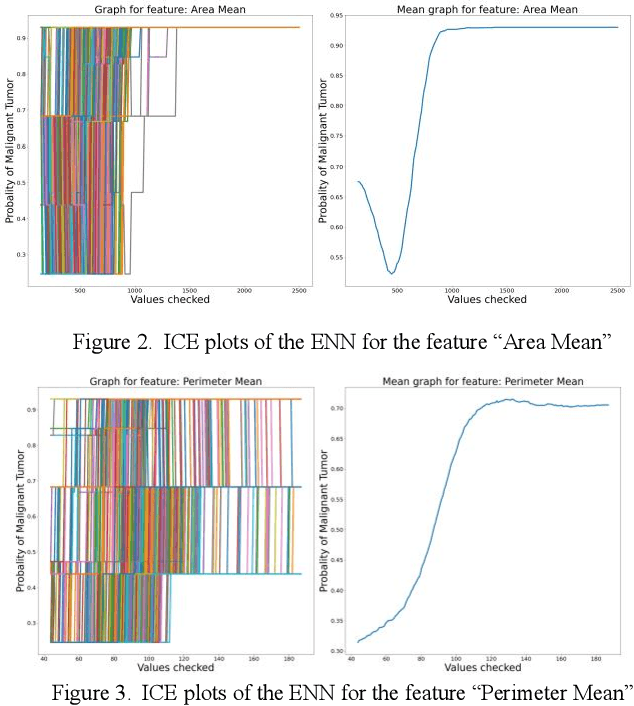
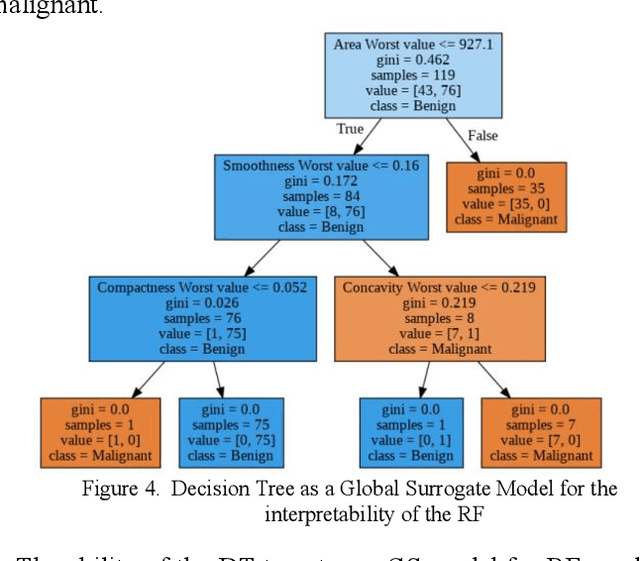
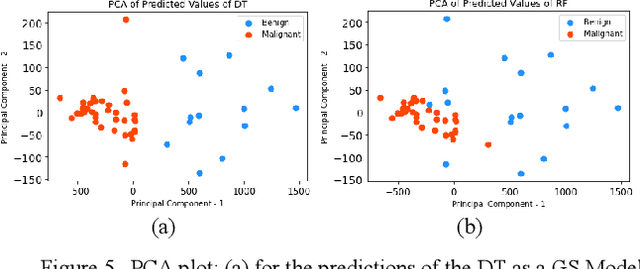
Abstract:Early detection of breast cancer is a powerful tool towards decreasing its socioeconomic burden. Although, artificial intelligence (AI) methods have shown remarkable results towards this goal, their "black box" nature hinders their wide adoption in clinical practice. To address the need for AI guided breast cancer diagnosis, interpretability methods can be utilized. In this study, we used AI methods, i.e., Random Forests (RF), Neural Networks (NN) and Ensembles of Neural Networks (ENN), towards this goal and explained and optimized their performance through interpretability techniques, such as the Global Surrogate (GS) method, the Individual Conditional Expectation (ICE) plots and the Shapley values (SV). The Wisconsin Diagnostic Breast Cancer (WDBC) dataset of the open UCI repository was used for the training and evaluation of the AI algorithms. The best performance for breast cancer diagnosis was achieved by the proposed ENN (96.6% accuracy and 0.96 area under the ROC curve), and its predictions were explained by ICE plots, proving that its decisions were compliant with current medical knowledge and can be further utilized to gain new insights in the pathophysiological mechanisms of breast cancer. Feature selection based on features' importance according to the GS model improved the performance of the RF (leading the accuracy from 96.49% to 97.18% and the area under the ROC curve from 0.96 to 0.97) and feature selection based on features' importance according to SV improved the performance of the NN (leading the accuracy from 94.6% to 95.53% and the area under the ROC curve from 0.94 to 0.95). Compared to other approaches on the same dataset, our proposed models demonstrated state of the art performance while being interpretable.
An explainable XGBoost-based approach towards assessing the risk of cardiovascular disease in patients with Type 2 Diabetes Mellitus
Sep 14, 2020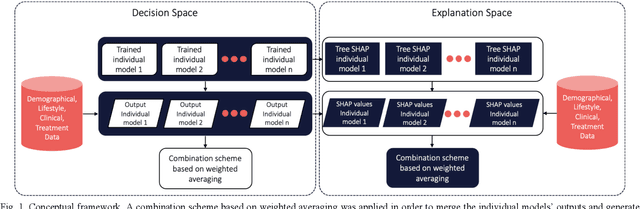
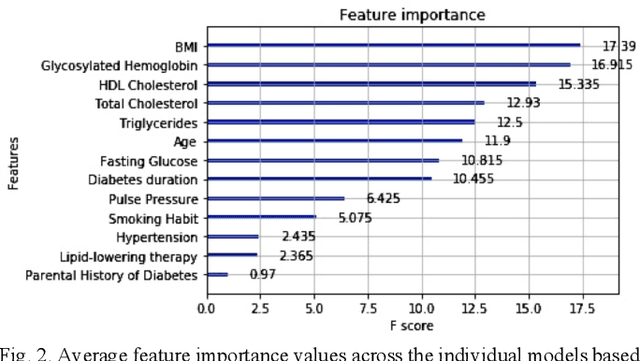
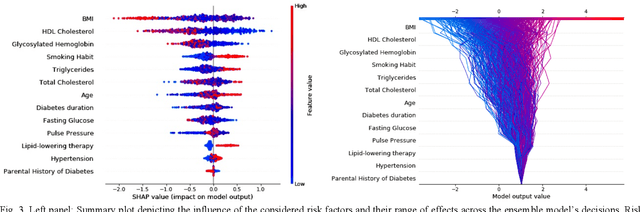
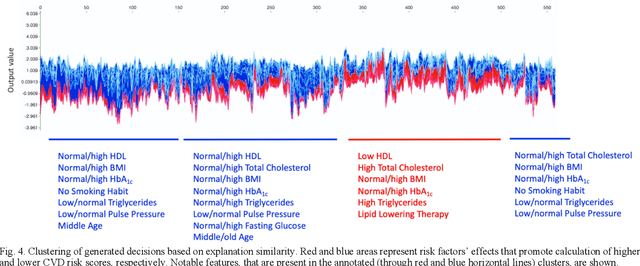
Abstract:Cardiovascular Disease (CVD) is an important cause of disability and death among individuals with Diabetes Mellitus (DM). International clinical guidelines for the management of Type 2 DM (T2DM) are founded on primary and secondary prevention and favor the evaluation of CVD related risk factors towards appropriate treatment initiation. CVD risk prediction models can provide valuable tools for optimizing the frequency of medical visits and performing timely preventive and therapeutic interventions against CVD events. The integration of explainability modalities in these models can enhance human understanding on the reasoning process, maximize transparency and embellish trust towards the models' adoption in clinical practice. The aim of the present study is to develop and evaluate an explainable personalized risk prediction model for the fatal or non-fatal CVD incidence in T2DM individuals. An explainable approach based on the eXtreme Gradient Boosting (XGBoost) and the Tree SHAP (SHapley Additive exPlanations) method is deployed for the calculation of the 5-year CVD risk and the generation of individual explanations on the model's decisions. Data from the 5-year follow up of 560 patients with T2DM are used for development and evaluation purposes. The obtained results (AUC = 71.13%) indicate the potential of the proposed approach to handle the unbalanced nature of the used dataset, while providing clinically meaningful insights about the ensemble model's decision process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge