Kira Radinsky
SFO: Learning PDE Operators via Spectral Filtering
Jan 23, 2026Abstract:Partial differential equations (PDEs) govern complex systems, yet neural operators often struggle to efficiently capture the long-range, nonlocal interactions inherent in their solution maps. We introduce Spectral Filtering Operator (SFO), a neural operator that parameterizes integral kernels using the Universal Spectral Basis (USB), a fixed, global orthonormal basis derived from the eigenmodes of the Hilbert matrix in spectral filtering theory. Motivated by our theoretical finding that the discrete Green's functions of shift-invariant PDE discretizations exhibit spatial Linear Dynamical System (LDS) structure, we prove that these kernels admit compact approximations in the USB. By learning only the spectral coefficients of rapidly decaying eigenvalues, SFO achieves a highly efficient representation. Across six benchmarks, including reaction-diffusion, fluid dynamics, and 3D electromagnetics, SFO achieves state-of-the-art accuracy, reducing error by up to 40% relative to strong baselines while using substantially fewer parameters.
SVD-NO: Learning PDE Solution Operators with SVD Integral Kernels
Nov 13, 2025Abstract:Neural operators have emerged as a promising paradigm for learning solution operators of partial differential equa- tions (PDEs) directly from data. Existing methods, such as those based on Fourier or graph techniques, make strong as- sumptions about the structure of the kernel integral opera- tor, assumptions which may limit expressivity. We present SVD-NO, a neural operator that explicitly parameterizes the kernel by its singular-value decomposition (SVD) and then carries out the integral directly in the low-rank basis. Two lightweight networks learn the left and right singular func- tions, a diagonal parameter matrix learns the singular values, and a Gram-matrix regularizer enforces orthonormality. As SVD-NO approximates the full kernel, it obtains a high de- gree of expressivity. Furthermore, due to its low-rank struc- ture the computational complexity of applying the operator remains reasonable, leading to a practical system. In exten- sive evaluations on five diverse benchmark equations, SVD- NO achieves a new state of the art. In particular, SVD-NO provides greater performance gains on PDEs whose solutions are highly spatially variable. The code of this work is publicly available at https://github.com/2noamk/SVDNO.git.
Docking-Aware Attention: Dynamic Protein Representations through Molecular Context Integration
Feb 03, 2025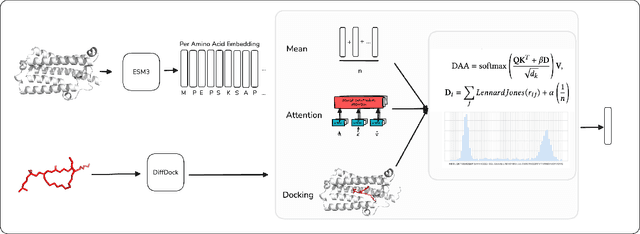
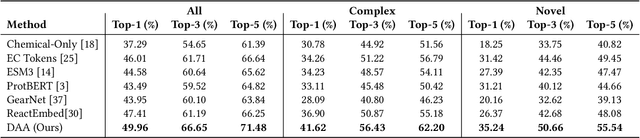
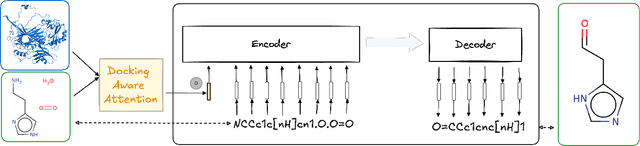
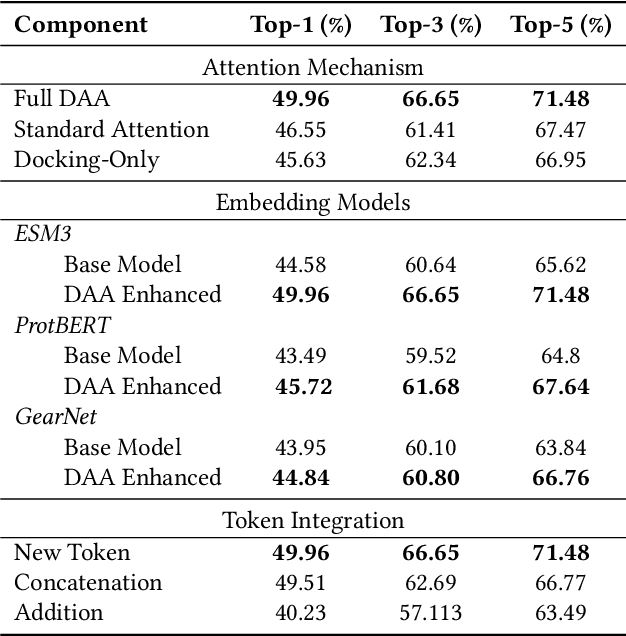
Abstract:Computational prediction of enzymatic reactions represents a crucial challenge in sustainable chemical synthesis across various scientific domains, ranging from drug discovery to materials science and green chemistry. These syntheses rely on proteins that selectively catalyze complex molecular transformations. These protein catalysts exhibit remarkable substrate adaptability, with the same protein often catalyzing different chemical transformations depending on its molecular partners. Current approaches to protein representation in reaction prediction either ignore protein structure entirely or rely on static embeddings, failing to capture how proteins dynamically adapt their behavior to different substrates. We present Docking-Aware Attention (DAA), a novel architecture that generates dynamic, context-dependent protein representations by incorporating molecular docking information into the attention mechanism. DAA combines physical interaction scores from docking predictions with learned attention patterns to focus on protein regions most relevant to specific molecular interactions. We evaluate our method on enzymatic reaction prediction, where it outperforms previous state-of-the-art methods, achieving 62.2\% accuracy versus 56.79\% on complex molecules and 55.54\% versus 49.45\% on innovative reactions. Through detailed ablation studies and visualizations, we demonstrate how DAA generates interpretable attention patterns that adapt to different molecular contexts. Our approach represents a general framework for context-aware protein representation in biocatalysis prediction, with potential applications across enzymatic synthesis planning. We open-source our implementation and pre-trained models to facilitate further research.
ReactEmbed: A Cross-Domain Framework for Protein-Molecule Representation Learning via Biochemical Reaction Networks
Jan 30, 2025Abstract:The challenge in computational biology and drug discovery lies in creating comprehensive representations of proteins and molecules that capture their intrinsic properties and interactions. Traditional methods often focus on unimodal data, such as protein sequences or molecular structures, limiting their ability to capture complex biochemical relationships. This work enhances these representations by integrating biochemical reactions encompassing interactions between molecules and proteins. By leveraging reaction data alongside pre-trained embeddings from state-of-the-art protein and molecule models, we develop ReactEmbed, a novel method that creates a unified embedding space through contrastive learning. We evaluate ReactEmbed across diverse tasks, including drug-target interaction, protein-protein interaction, protein property prediction, and molecular property prediction, consistently surpassing all current state-of-the-art models. Notably, we showcase ReactEmbed's practical utility through successful implementation in lipid nanoparticle-based drug delivery, enabling zero-shot prediction of blood-brain barrier permeability for protein-nanoparticle complexes. The code and comprehensive database of reaction pairs are available for open use at \href{https://github.com/amitaysicherman/ReactEmbed}{GitHub}.
Ordinary Differential Equations for Enhanced 12-Lead ECG Generation
Sep 26, 2024



Abstract:In the realm of artificial intelligence, the generation of realistic training data for supervised learning tasks presents a significant challenge. This is particularly true in the synthesis of electrocardiograms (ECGs), where the objective is to develop a synthetic 12-lead ECG model. The primary complexity of this task stems from accurately modeling the intricate biological and physiological interactions among different ECG leads. Although mathematical process simulators have shed light on these dynamics, effectively incorporating this understanding into generative models is not straightforward. In this work, we introduce an innovative method that employs ordinary differential equations (ODEs) to enhance the fidelity of generating 12-lead ECG data. This approach integrates a system of ODEs that represent cardiac dynamics directly into the generative model's optimization process, allowing for the production of biologically plausible ECG training data that authentically reflects real-world variability and inter-lead dependencies. We conducted an empirical analysis of thousands of ECGs and found that incorporating cardiac simulation insights into the data generation process significantly improves the accuracy of heart abnormality classifiers trained on this synthetic 12-lead ECG data.
GOProteinGNN: Leveraging Protein Knowledge Graphs for Protein Representation Learning
Jul 31, 2024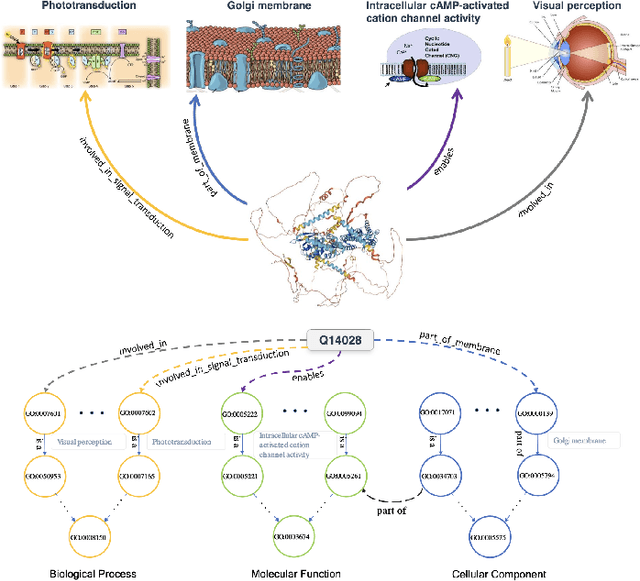
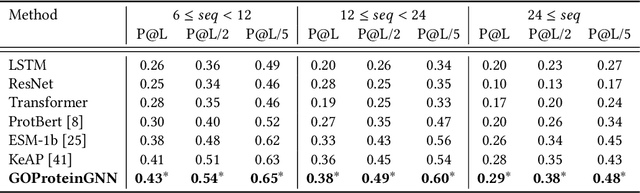
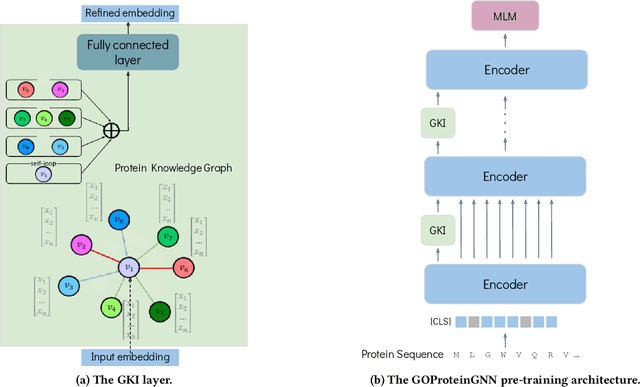
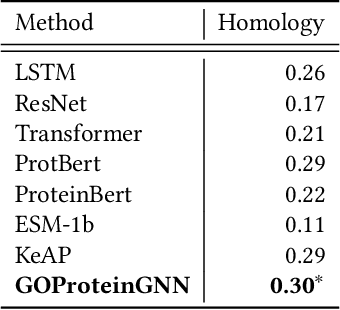
Abstract:Proteins play a vital role in biological processes and are indispensable for living organisms. Accurate representation of proteins is crucial, especially in drug development. Recently, there has been a notable increase in interest in utilizing machine learning and deep learning techniques for unsupervised learning of protein representations. However, these approaches often focus solely on the amino acid sequence of proteins and lack factual knowledge about proteins and their interactions, thus limiting their performance. In this study, we present GOProteinGNN, a novel architecture that enhances protein language models by integrating protein knowledge graph information during the creation of amino acid level representations. Our approach allows for the integration of information at both the individual amino acid level and the entire protein level, enabling a comprehensive and effective learning process through graph-based learning. By doing so, we can capture complex relationships and dependencies between proteins and their functional annotations, resulting in more robust and contextually enriched protein representations. Unlike previous fusion methods, GOProteinGNN uniquely learns the entire protein knowledge graph during training, which allows it to capture broader relational nuances and dependencies beyond mere triplets as done in previous work. We perform a comprehensive evaluation on several downstream tasks demonstrating that GOProteinGNN consistently outperforms previous methods, showcasing its effectiveness and establishing it as a state-of-the-art solution for protein representation learning.
Interpretable Multivariate Time Series Forecasting Using Neural Fourier Transform
May 22, 2024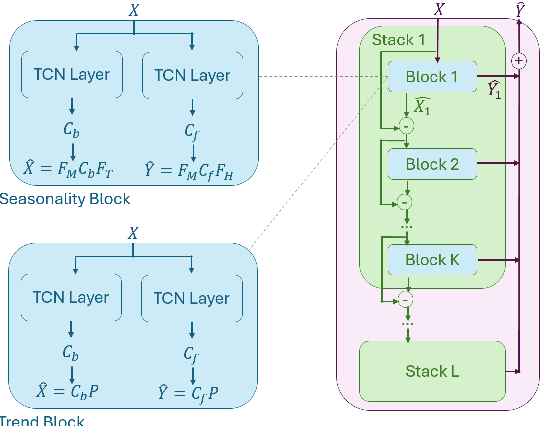
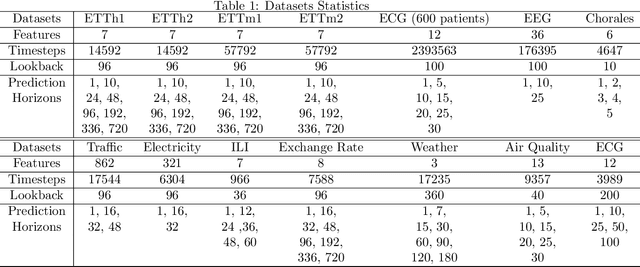
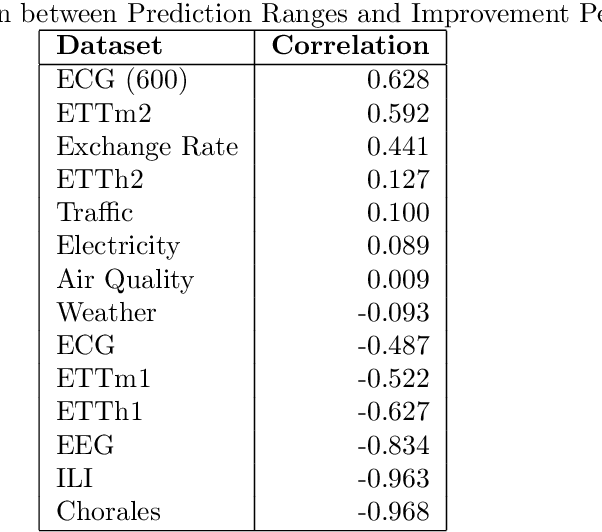
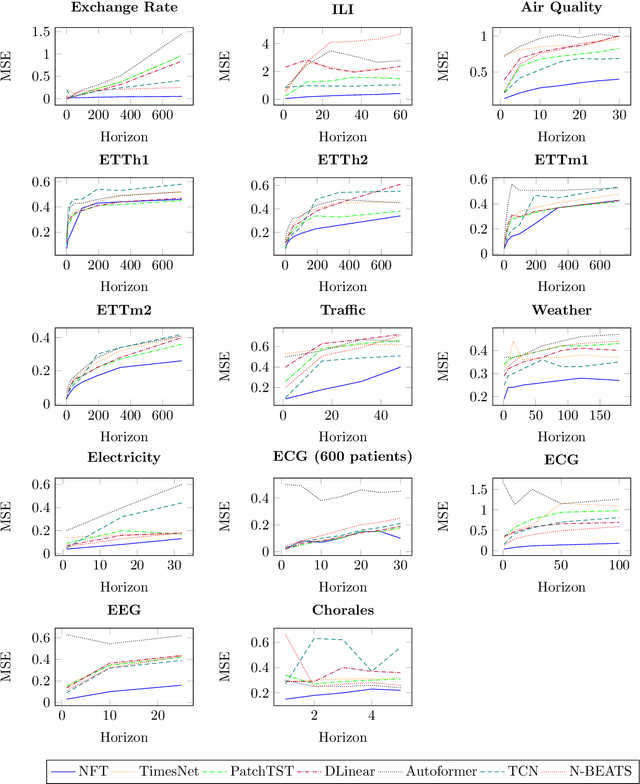
Abstract:Multivariate time series forecasting is a pivotal task in several domains, including financial planning, medical diagnostics, and climate science. This paper presents the Neural Fourier Transform (NFT) algorithm, which combines multi-dimensional Fourier transforms with Temporal Convolutional Network layers to improve both the accuracy and interpretability of forecasts. The Neural Fourier Transform is empirically validated on fourteen diverse datasets, showing superior performance across multiple forecasting horizons and lookbacks, setting new benchmarks in the field. This work advances multivariate time series forecasting by providing a model that is both interpretable and highly predictive, making it a valuable tool for both practitioners and researchers. The code for this study is publicly available.
Leveraging World Events to Predict E-Commerce Consumer Demand under Anomaly
May 22, 2024Abstract:Consumer demand forecasting is of high importance for many e-commerce applications, including supply chain optimization, advertisement placement, and delivery speed optimization. However, reliable time series sales forecasting for e-commerce is difficult, especially during periods with many anomalies, as can often happen during pandemics, abnormal weather, or sports events. Although many time series algorithms have been applied to the task, prediction during anomalies still remains a challenge. In this work, we hypothesize that leveraging external knowledge found in world events can help overcome the challenge of prediction under anomalies. We mine a large repository of 40 years of world events and their textual representations. Further, we present a novel methodology based on transformers to construct an embedding of a day based on the relations of the day's events. Those embeddings are then used to forecast future consumer behavior. We empirically evaluate the methods over a large e-commerce products sales dataset, extracted from eBay, one of the world's largest online marketplaces. We show over numerous categories that our method outperforms state-of-the-art baselines during anomalies.
Leveraging Prototypical Representations for Mitigating Social Bias without Demographic Information
Mar 14, 2024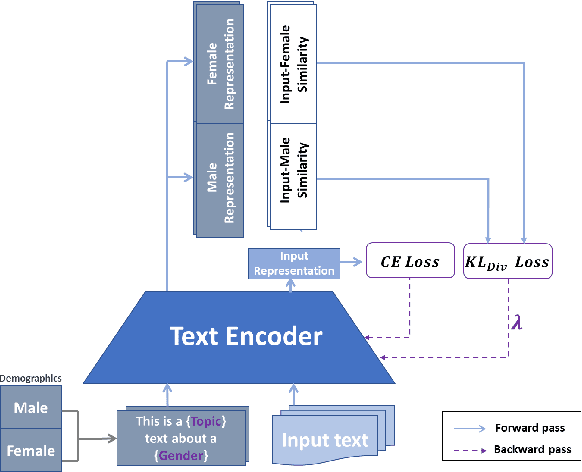
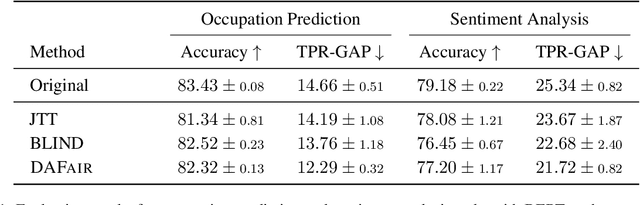
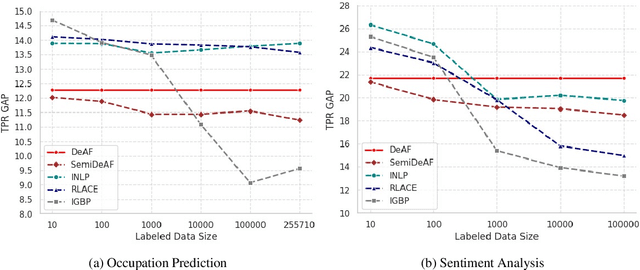
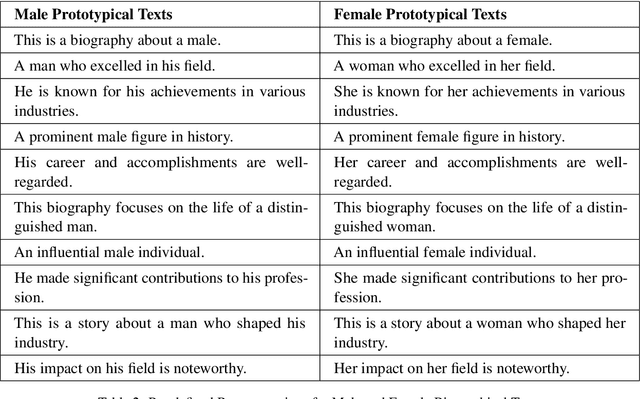
Abstract:Mitigating social biases typically requires identifying the social groups associated with each data sample. In this paper, we present DAFair, a novel approach to address social bias in language models. Unlike traditional methods that rely on explicit demographic labels, our approach does not require any such information. Instead, we leverage predefined prototypical demographic texts and incorporate a regularization term during the fine-tuning process to mitigate bias in the model's representations. Our empirical results across two tasks and two models demonstrate the effectiveness of our method compared to previous approaches that do not rely on labeled data. Moreover, with limited demographic-annotated data, our approach outperforms common debiasing approaches.
Shielded Representations: Protecting Sensitive Attributes Through Iterative Gradient-Based Projection
May 17, 2023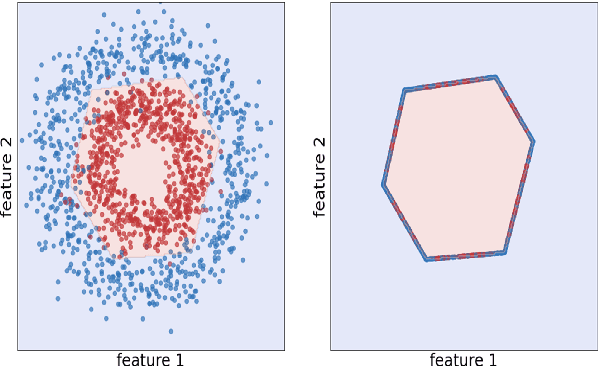
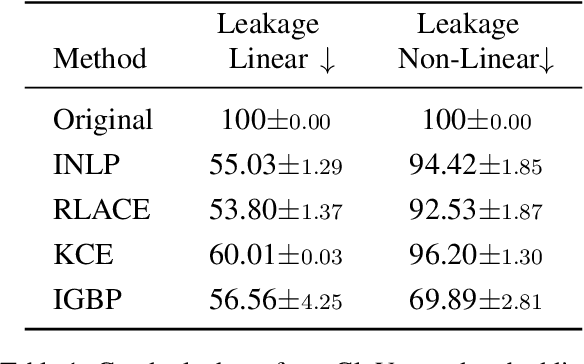
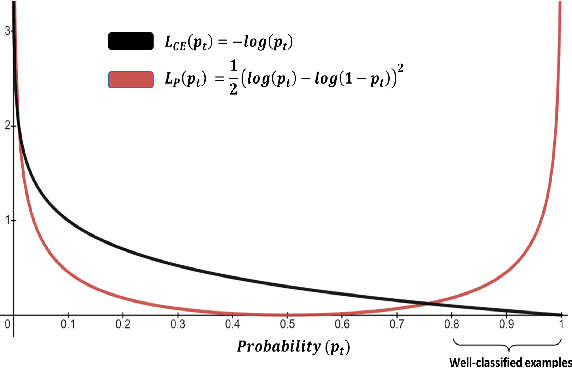
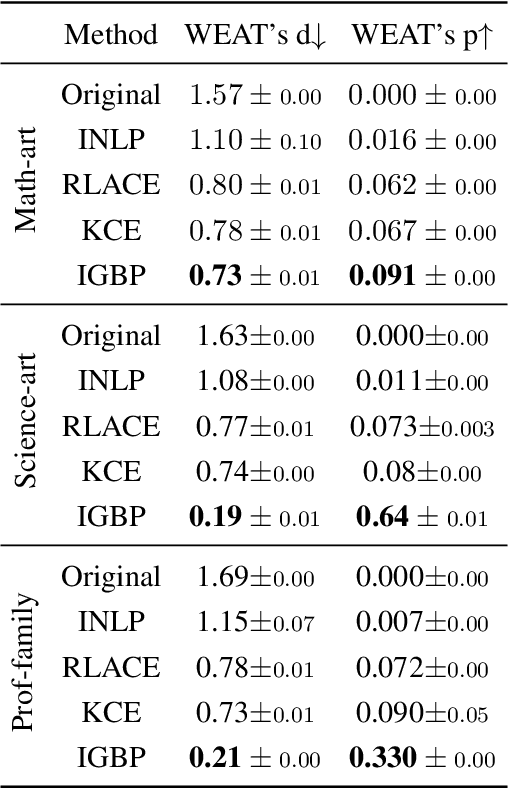
Abstract:Natural language processing models tend to learn and encode social biases present in the data. One popular approach for addressing such biases is to eliminate encoded information from the model's representations. However, current methods are restricted to removing only linearly encoded information. In this work, we propose Iterative Gradient-Based Projection (IGBP), a novel method for removing non-linear encoded concepts from neural representations. Our method consists of iteratively training neural classifiers to predict a particular attribute we seek to eliminate, followed by a projection of the representation on a hypersurface, such that the classifiers become oblivious to the target attribute. We evaluate the effectiveness of our method on the task of removing gender and race information as sensitive attributes. Our results demonstrate that IGBP is effective in mitigating bias through intrinsic and extrinsic evaluations, with minimal impact on downstream task accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge