Amitay Sicherman
Docking-Aware Attention: Dynamic Protein Representations through Molecular Context Integration
Feb 03, 2025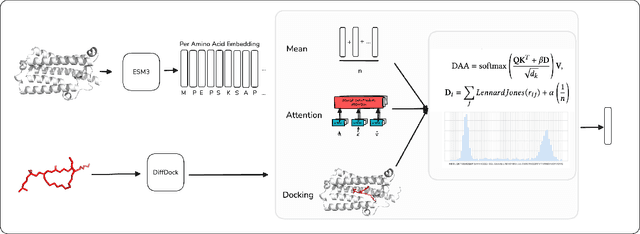
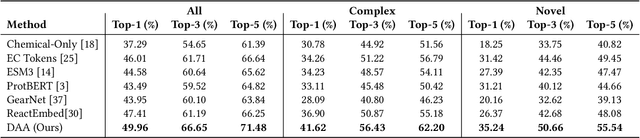
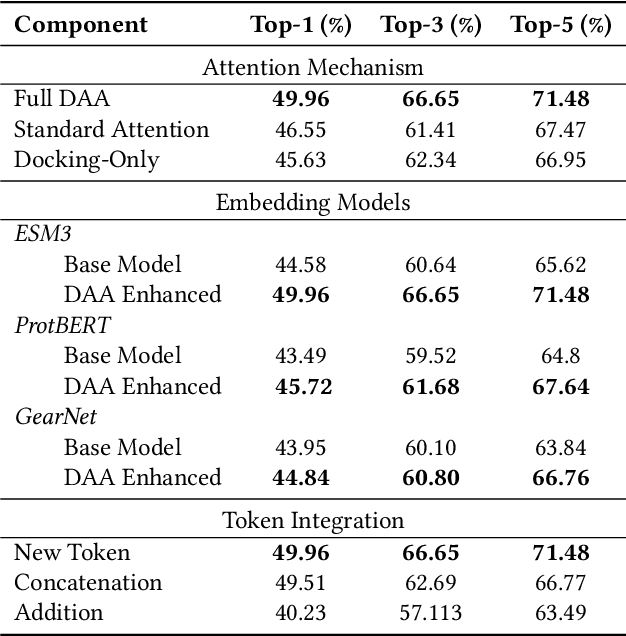
Abstract:Computational prediction of enzymatic reactions represents a crucial challenge in sustainable chemical synthesis across various scientific domains, ranging from drug discovery to materials science and green chemistry. These syntheses rely on proteins that selectively catalyze complex molecular transformations. These protein catalysts exhibit remarkable substrate adaptability, with the same protein often catalyzing different chemical transformations depending on its molecular partners. Current approaches to protein representation in reaction prediction either ignore protein structure entirely or rely on static embeddings, failing to capture how proteins dynamically adapt their behavior to different substrates. We present Docking-Aware Attention (DAA), a novel architecture that generates dynamic, context-dependent protein representations by incorporating molecular docking information into the attention mechanism. DAA combines physical interaction scores from docking predictions with learned attention patterns to focus on protein regions most relevant to specific molecular interactions. We evaluate our method on enzymatic reaction prediction, where it outperforms previous state-of-the-art methods, achieving 62.2\% accuracy versus 56.79\% on complex molecules and 55.54\% versus 49.45\% on innovative reactions. Through detailed ablation studies and visualizations, we demonstrate how DAA generates interpretable attention patterns that adapt to different molecular contexts. Our approach represents a general framework for context-aware protein representation in biocatalysis prediction, with potential applications across enzymatic synthesis planning. We open-source our implementation and pre-trained models to facilitate further research.
ReactEmbed: A Cross-Domain Framework for Protein-Molecule Representation Learning via Biochemical Reaction Networks
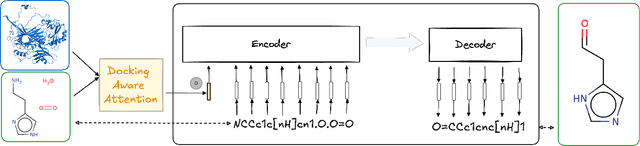
Jan 30, 2025Abstract:The challenge in computational biology and drug discovery lies in creating comprehensive representations of proteins and molecules that capture their intrinsic properties and interactions. Traditional methods often focus on unimodal data, such as protein sequences or molecular structures, limiting their ability to capture complex biochemical relationships. This work enhances these representations by integrating biochemical reactions encompassing interactions between molecules and proteins. By leveraging reaction data alongside pre-trained embeddings from state-of-the-art protein and molecule models, we develop ReactEmbed, a novel method that creates a unified embedding space through contrastive learning. We evaluate ReactEmbed across diverse tasks, including drug-target interaction, protein-protein interaction, protein property prediction, and molecular property prediction, consistently surpassing all current state-of-the-art models. Notably, we showcase ReactEmbed's practical utility through successful implementation in lipid nanoparticle-based drug delivery, enabling zero-shot prediction of blood-brain barrier permeability for protein-nanoparticle complexes. The code and comprehensive database of reaction pairs are available for open use at \href{https://github.com/amitaysicherman/ReactEmbed}{GitHub}.
Analysing Discrete Self Supervised Speech Representation for Spoken Language Modeling
Jan 02, 2023Abstract:This work profoundly analyzes discrete self-supervised speech representations through the eyes of Generative Spoken Language Modeling (GSLM). Following the findings of such an analysis, we propose practical improvements to the discrete unit for the GSLM. First, we start comprehending these units by analyzing them in three axes: interpretation, visualization, and resynthesis. Our analysis finds a high correlation between the speech units to phonemes and phoneme families, while their correlation with speaker or gender is weaker. Additionally, we found redundancies in the extracted units and claim that one reason may be the units' context. Following this analysis, we propose a new, unsupervised metric to measure unit redundancies. Finally, we use this metric to develop new methods that improve the robustness of units clustering and show significant improvement considering zero-resource speech metrics such as ABX. Code and analysis tools are available under the following link.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge