GOProteinGNN: Leveraging Protein Knowledge Graphs for Protein Representation Learning
Paper and Code
Jul 31, 2024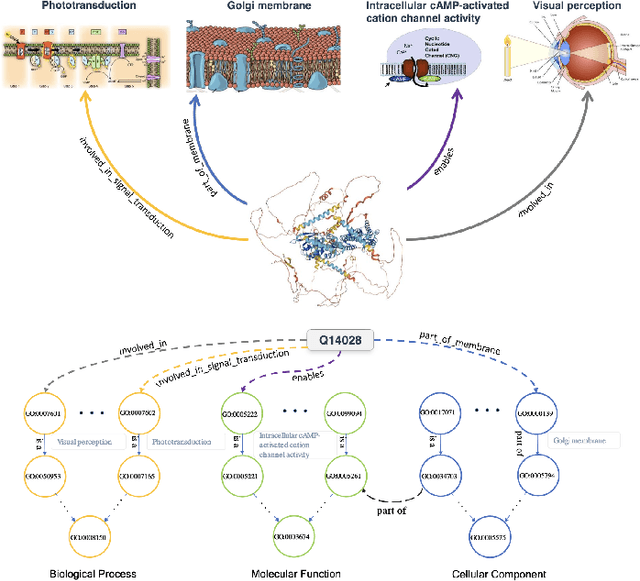
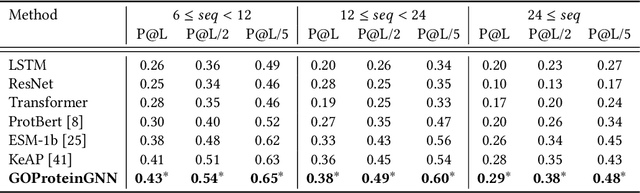
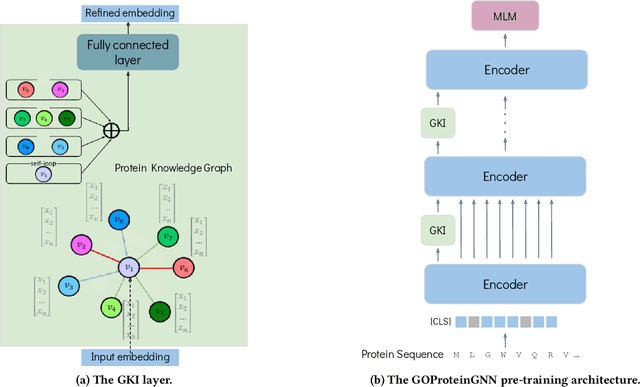
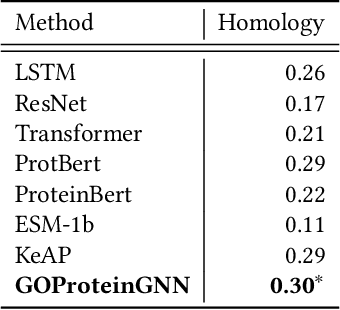
Proteins play a vital role in biological processes and are indispensable for living organisms. Accurate representation of proteins is crucial, especially in drug development. Recently, there has been a notable increase in interest in utilizing machine learning and deep learning techniques for unsupervised learning of protein representations. However, these approaches often focus solely on the amino acid sequence of proteins and lack factual knowledge about proteins and their interactions, thus limiting their performance. In this study, we present GOProteinGNN, a novel architecture that enhances protein language models by integrating protein knowledge graph information during the creation of amino acid level representations. Our approach allows for the integration of information at both the individual amino acid level and the entire protein level, enabling a comprehensive and effective learning process through graph-based learning. By doing so, we can capture complex relationships and dependencies between proteins and their functional annotations, resulting in more robust and contextually enriched protein representations. Unlike previous fusion methods, GOProteinGNN uniquely learns the entire protein knowledge graph during training, which allows it to capture broader relational nuances and dependencies beyond mere triplets as done in previous work. We perform a comprehensive evaluation on several downstream tasks demonstrating that GOProteinGNN consistently outperforms previous methods, showcasing its effectiveness and establishing it as a state-of-the-art solution for protein representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge