Kevin Dhaliwal
ROOM: A Physics-Based Continuum Robot Simulator for Photorealistic Medical Datasets Generation
Sep 16, 2025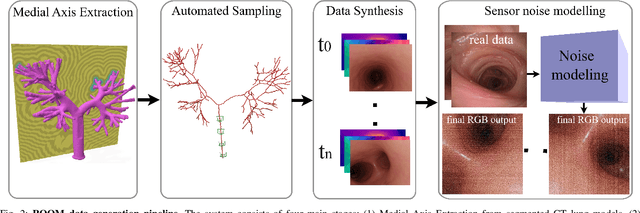



Abstract:Continuum robots are advancing bronchoscopy procedures by accessing complex lung airways and enabling targeted interventions. However, their development is limited by the lack of realistic training and test environments: Real data is difficult to collect due to ethical constraints and patient safety concerns, and developing autonomy algorithms requires realistic imaging and physical feedback. We present ROOM (Realistic Optical Observation in Medicine), a comprehensive simulation framework designed for generating photorealistic bronchoscopy training data. By leveraging patient CT scans, our pipeline renders multi-modal sensor data including RGB images with realistic noise and light specularities, metric depth maps, surface normals, optical flow and point clouds at medically relevant scales. We validate the data generated by ROOM in two canonical tasks for medical robotics -- multi-view pose estimation and monocular depth estimation, demonstrating diverse challenges that state-of-the-art methods must overcome to transfer to these medical settings. Furthermore, we show that the data produced by ROOM can be used to fine-tune existing depth estimation models to overcome these challenges, also enabling other downstream applications such as navigation. We expect that ROOM will enable large-scale data generation across diverse patient anatomies and procedural scenarios that are challenging to capture in clinical settings. Code and data: https://github.com/iamsalvatore/room.
Deep Learning-Assisted Co-registration of Full-Spectral Autofluorescence Lifetime Microscopic Images with H&E-Stained Histology Images
Feb 15, 2022



Abstract:Autofluorescence lifetime images reveal unique characteristics of endogenous fluorescence in biological samples. Comprehensive understanding and clinical diagnosis rely on co-registration with the gold standard, histology images, which is extremely challenging due to the difference of both images. Here, we show an unsupervised image-to-image translation network that significantly improves the success of the co-registration using a conventional optimisation-based regression network, applicable to autofluorescence lifetime images at different emission wavelengths. A preliminary blind comparison by experienced researchers shows the superiority of our method on co-registration. The results also indicate that the approach is applicable to various image formats, like fluorescence intensity images. With the registration, stitching outcomes illustrate the distinct differences of the spectral lifetime across an unstained tissue, enabling macro-level rapid visual identification of lung cancer and cellular-level characterisation of cell variants and common types. The approach could be effortlessly extended to lifetime images beyond this range and other staining technologies.
Patch-based Sparse Representation For Bacterial Detection
Oct 29, 2018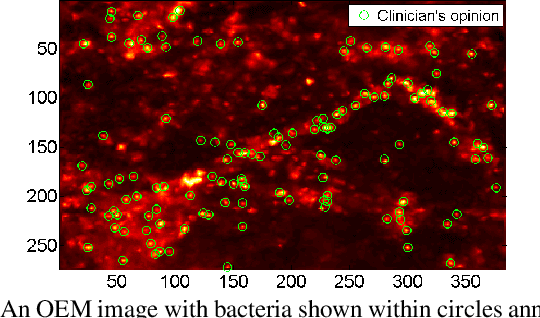
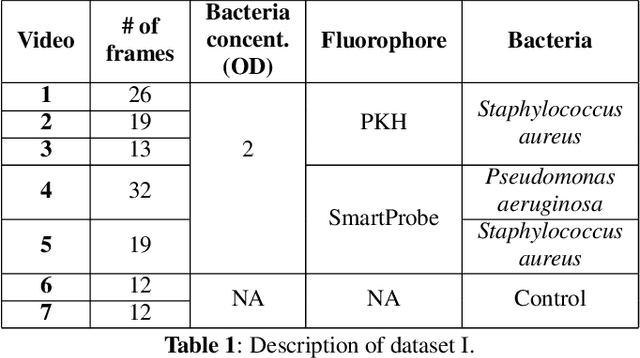
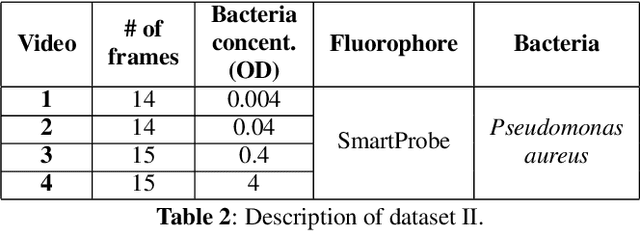
Abstract:In this paper, we propose a supervised approach for bacterial detection in optical endomicroscopy images. This approach splits each image into a set of overlapping patches and assumes that observed intensities are linear combinations of the actual intensity values associated with background image structures, corrupted by additive Gaussian noise and potentially by a sparse outlier term modelling anomalies (which are considered to be candidate bacteria). The actual intensity term representing background structures is modelled as a linear combination of a few atoms drawn from a dictionary which is learned from bacteria-free data and then fixed while analyzing new images. The bacteria detection task is formulated as a minimization problem and an Alternating Direction Method of Multipliers (ADMM) is then used to estimate the unknown parameters. Simulations conducted using two ex vivo lung datasets show good detection and correlation performance between bacteria counts identified by a trained clinician and those of the proposed method.
Image computing for fibre-bundle endomicroscopy: A review
Sep 03, 2018



Abstract:Endomicroscopy is an emerging imaging modality, that facilitates the acquisition of in vivo, in situ optical biopsies, assisting diagnostic and potentially therapeutic interventions. While there is a diverse and constantly expanding range of commercial and experimental optical biopsy platforms available, fibre-bundle endomicroscopy is currently the most widely used platform and is approved for clinical use in a range of clinical indications. Miniaturised, flexible fibre-bundles, guided through the working channel of endoscopes, needles and catheters, enable high-resolution imaging across a variety of organ systems. Yet, the nature of image acquisition though a fibre-bundle gives rise to several inherent characteristics and limitations necessitating novel and effective image pre- and post-processing algorithms, ranging from image formation, enhancement and mosaicing to pathology detection and quantification. This paper introduces the underlying technology and most prevalent clinical applications of fibre-bundle endomicroscopy, and provides a comprehensive, up-to-date, review of relevant image reconstruction, analysis and understanding/inference methodologies. Furthermore, current limitations as well as future challenges and opportunities in fibre-bundle endomicroscopy computing are identified and discussed.
Deconvolution and Restoration of Optical Endomicroscopy Images
Aug 28, 2018



Abstract:Optical endomicroscopy (OEM) is an emerging technology platform with preclinical and clinical imaging applications. Pulmonary OEM via fibre bundles has the potential to provide in vivo, in situ molecular signatures of disease such as infection and inflammation. However, enhancing the quality of data acquired by this technique for better visualization and subsequent analysis remains a challenging problem. Cross coupling between fiber cores and sparse sampling by imaging fiber bundles are the main reasons for image degradation, and poor detection performance (i.e., inflammation, bacteria, etc.). In this work, we address the problem of deconvolution and restoration of OEM data. We propose a hierarchical Bayesian model to solve this problem and compare three estimation algorithms to exploit the resulting joint posterior distribution. The first method is based on Markov chain Monte Carlo (MCMC) methods, however, it exhibits a relatively long computational time. The second and third algorithms deal with this issue and are based on a variational Bayes (VB) approach and an alternating direction method of multipliers (ADMM) algorithm respectively. Results on both synthetic and real datasets illustrate the effectiveness of the proposed methods for restoration of OEM images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge