Kevin Brown
The Patient is not a Moving Document: A World Model Training Paradigm for Longitudinal EHR
Jan 29, 2026Abstract:Large language models (LLMs) trained with next-word-prediction have achieved success as clinical foundation models. Representations from these language backbones yield strong linear probe performance across biomedical tasks, suggesting that patient semantics emerge from next-token prediction at scale. However, this paradigm treats patients as a document to be summarized rather than a dynamical system to be simulated; a patient's trajectory emerges from their state evolving under interventions and time, requiring models that simulate dynamics rather than predict tokens. To address this, we introduce SMB-Structure, a world model for structured EHR that grounds a joint-embedding prediction architecture (JEPA) with next-token prediction (SFT). SFT grounds our model to reconstruct future patient states in token space, while JEPA predicts those futures in latent space from the initial patient representation alone, forcing trajectory dynamics to be encoded before the next state is observed. We validate across two large-scale cohorts: Memorial Sloan Kettering (23,319 oncology patients; 323,000+ patient-years) and INSPECT (19,402 pulmonary embolism patients). Using a linear probe evaluated at multiple points along the disease trajectory, we demonstrate that our training paradigm learns embeddings that capture disease dynamics not recoverable by autoregressive baselines, enabling SMB-Structure to achieve competitive performance on complex tasks characterized by high patient heterogeneity. Model weights are available at https://huggingface.co/standardmodelbio/SMB-v1-1.7B-Structure.
Advancing High Resolution Vision-Language Models in Biomedicine
Jun 12, 2024Abstract:Multi-modal learning has significantly advanced generative AI, especially in vision-language modeling. Innovations like GPT-4V and open-source projects such as LLaVA have enabled robust conversational agents capable of zero-shot task completions. However, applying these technologies in the biomedical field presents unique challenges. Recent initiatives like LLaVA-Med have started to adapt instruction-tuning for biomedical contexts using large datasets such as PMC-15M. Our research offers three key contributions: (i) we present a new instruct dataset enriched with medical image-text pairs from Claude3-Opus and LLaMA3 70B, (ii) we propose a novel image encoding strategy using hierarchical representations to improve fine-grained biomedical visual comprehension, and (iii) we develop the Llama3-Med model, which achieves state-of-the-art zero-shot performance on biomedical visual question answering benchmarks, with an average performance improvement of over 10% compared to previous methods. These advancements provide more accurate and reliable tools for medical professionals, bridging gaps in current multi-modal conversational assistants and promoting further innovations in medical AI.
Masked Image Modeling Advances 3D Medical Image Analysis
Apr 25, 2022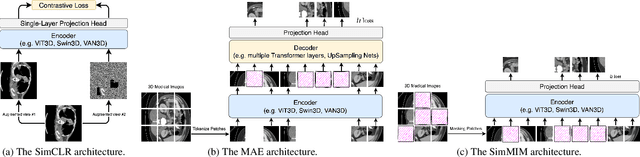
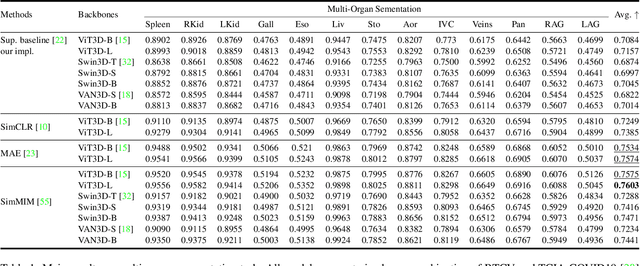
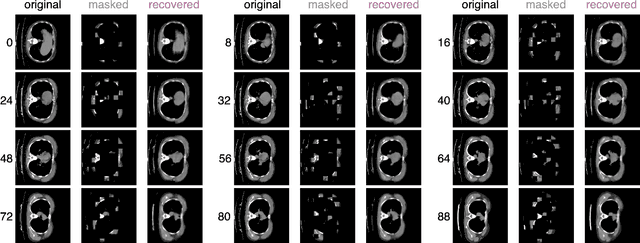
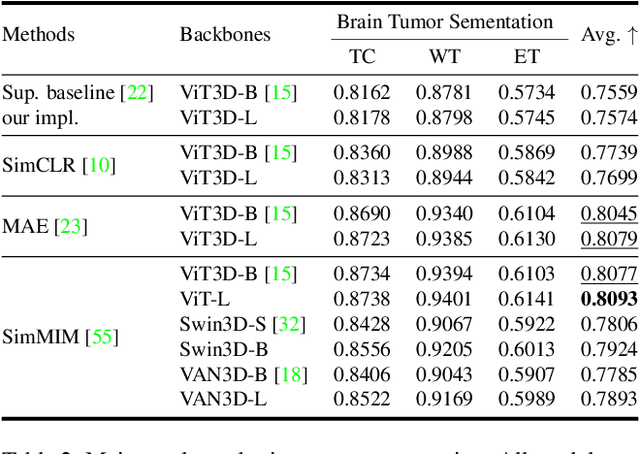
Abstract:Recently, masked image modeling (MIM) has gained considerable attention due to its capacity to learn from vast amounts of unlabeled data and has been demonstrated to be effective on a wide variety of vision tasks involving natural images. Meanwhile, the potential of self-supervised learning in modeling 3D medical images is anticipated to be immense due to the high quantities of unlabeled images, and the expense and difficulty of quality labels. However, MIM's applicability to medical images remains uncertain. In this paper, we demonstrate that masked image modeling approaches can also advance 3D medical images analysis in addition to natural images. We study how masked image modeling strategies leverage performance from the viewpoints of 3D medical image segmentation as a representative downstream task: i) when compared to naive contrastive learning, masked image modeling approaches accelerate the convergence of supervised training even faster (1.40$\times$) and ultimately produce a higher dice score; ii) predicting raw voxel values with a high masking ratio and a relatively smaller patch size is non-trivial self-supervised pretext-task for medical images modeling; iii) a lightweight decoder or projection head design for reconstruction is powerful for masked image modeling on 3D medical images which speeds up training and reduce cost; iv) finally, we also investigate the effectiveness of MIM methods under different practical scenarios where different image resolutions and labeled data ratios are applied.
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020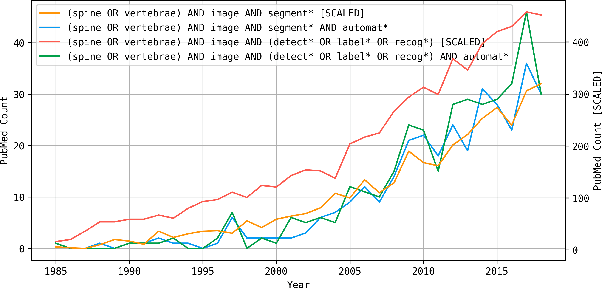
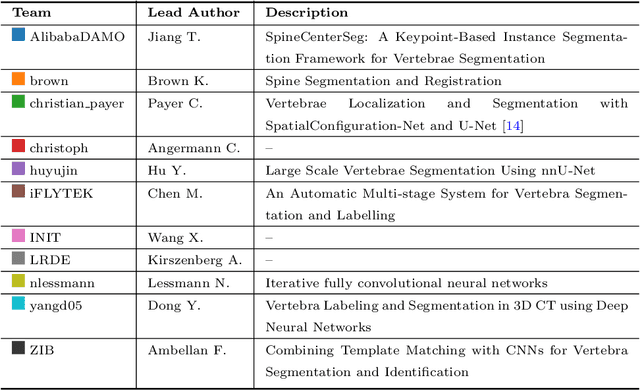
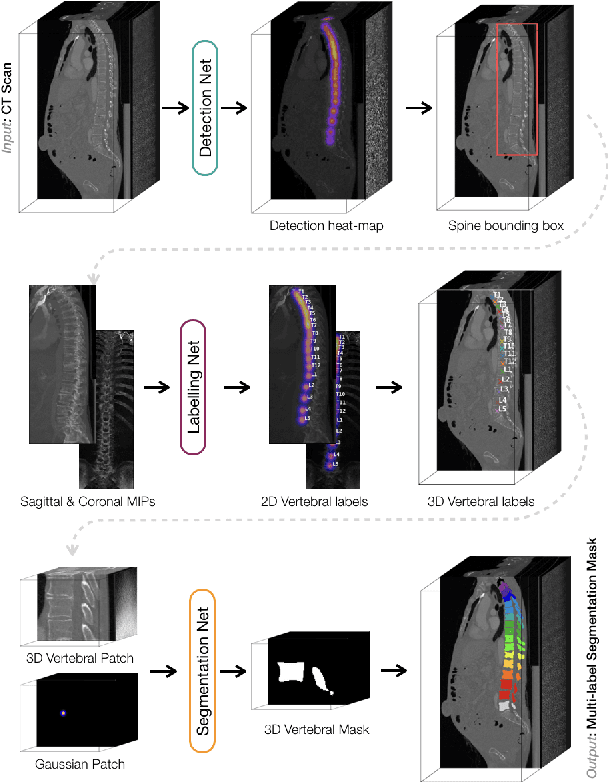
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge