Kei Sano
Data downlink prioritization using image classification on-board a 6U CubeSat
Aug 27, 2024Abstract:Nanosatellites are proliferating as low-cost dedicated sensing systems with lean development cycles. Kyushu Institute of Technology and collaborators have launched a joint venture for a nanosatellite mission, VERTECS. The primary mission is to elucidate the formation history of stars by observing the optical-wavelength cosmic background radiation. The VERTECS satellite will be equipped with a small-aperture telescope and a high-precision attitude control system to capture the cosmic data for analysis on the ground. However, nanosatellites are limited by their onboard memory resources and downlink speed capabilities. Additionally, due to a limited number of ground stations, the satellite mission will face issues meeting the required data budget for mission success. To alleviate this issue, we propose an on-orbit system to autonomously classify and then compress desirable image data for data downlink prioritization and optimization. The system comprises a prototype Camera Controller Board (CCB) which carries a Raspberry Pi Compute Module 4 which is used for classification and compression. The system uses a lightweight Convolutional Neural Network (CNN) model to classify and determine the desirability of captured image data. The model is designed to be lean and robust to reduce the computational and memory load on the satellite. The model is trained and tested on a novel star field dataset consisting of data captured by the Sloan Digital Sky Survey (SDSS). The dataset is meant to simulate the expected data produced by the 6U satellite. The compression step implements GZip, RICE or HCOMPRESS compression, which are standards for astronomical data. Preliminary testing on the proposed CNN model results in a classification accuracy of about 100\% on the star field dataset, with compression ratios of 3.99, 5.16 and 5.43 for GZip, RICE and HCOMPRESS that were achieved on tested FITS image data.
* 14 pages
Spatio-temporal reconstruction of substance dynamics using compressed sensing in multi-spectral magnetic resonance spectroscopic imaging
Mar 01, 2024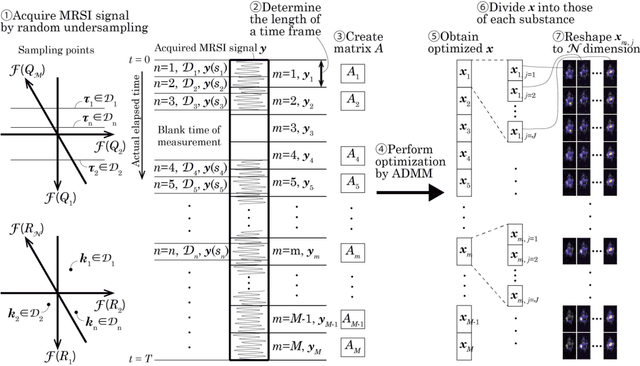
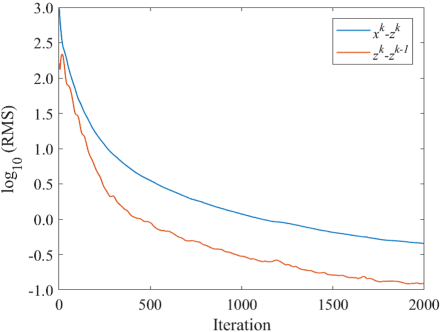
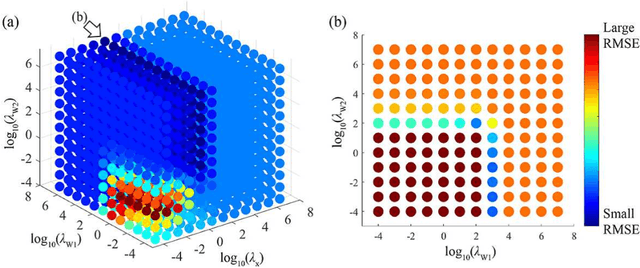
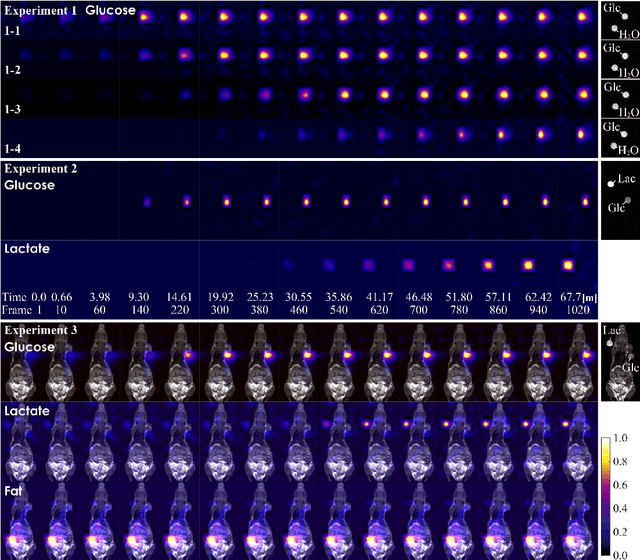
Abstract:The objective of our study is to observe dynamics of multiple substances in vivo with high temporal resolution from multi-spectral magnetic resonance spectroscopic imaging (MRSI) data. The multi-spectral MRSI can effectively separate spectral peaks of multiple substances and is useful to measure spatial distributions of substances. However it is difficult to measure time-varying substance distributions directly by ordinary full sampling because the measurement requires a significantly long time. In this study, we propose a novel method to reconstruct the spatio-temporal distributions of substances from randomly undersampled multi-spectral MRSI data on the basis of compressed sensing (CS) and the partially separable function model with base spectra of substances. In our method, we have employed spatio-temporal sparsity and temporal smoothness of the substance distributions as prior knowledge to perform CS. The effectiveness of our method has been evaluated using phantom data sets of glass tubes filled with glucose or lactate solution in increasing amounts over time and animal data sets of a tumor-bearing mouse to observe the metabolic dynamics involved in the Warburg effect in vivo. The reconstructed results are consistent with the expected behaviors, showing that our method can reconstruct the spatio-temporal distribution of substances with a temporal resolution of four seconds which is extremely short time scale compared with that of full sampling. Since this method utilizes only prior knowledge naturally assumed for the spatio-temporal distributions of substances and is independent of the number of the spectral and spatial dimensions or the acquisition sequence of MRSI, it is expected to contribute to revealing the underlying substance dynamics in MRSI data already acquired or to be acquired in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge