Katja Herzog
Grouping Shapley Value Feature Importances of Random Forests for explainable Yield Prediction
Apr 14, 2023Abstract:Explainability in yield prediction helps us fully explore the potential of machine learning models that are already able to achieve high accuracy for a variety of yield prediction scenarios. The data included for the prediction of yields are intricate and the models are often difficult to understand. However, understanding the models can be simplified by using natural groupings of the input features. Grouping can be achieved, for example, by the time the features are captured or by the sensor used to do so. The state-of-the-art for interpreting machine learning models is currently defined by the game-theoretic approach of Shapley values. To handle groups of features, the calculated Shapley values are typically added together, ignoring the theoretical limitations of this approach. We explain the concept of Shapley values directly computed for predefined groups of features and introduce an algorithm to compute them efficiently on tree structures. We provide a blueprint for designing swarm plots that combine many local explanations for global understanding. Extensive evaluation of two different yield prediction problems shows the worth of our approach and demonstrates how we can enable a better understanding of yield prediction models in the future, ultimately leading to mutual enrichment of research and application.
An Adaptive Approach for Automated Grapevine Phenotyping using VGG-based Convolutional Neural Networks
Nov 23, 2018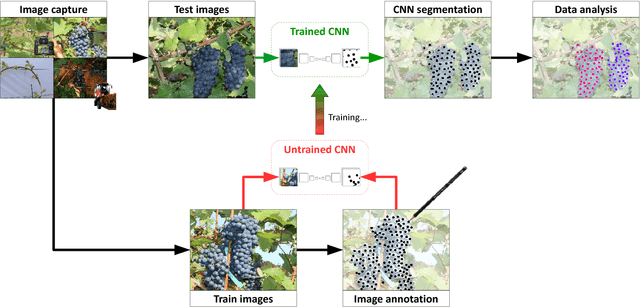
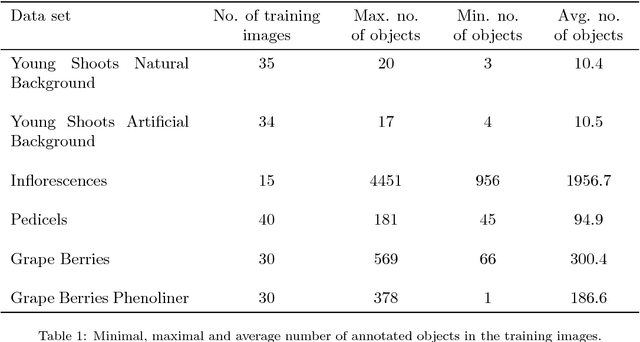
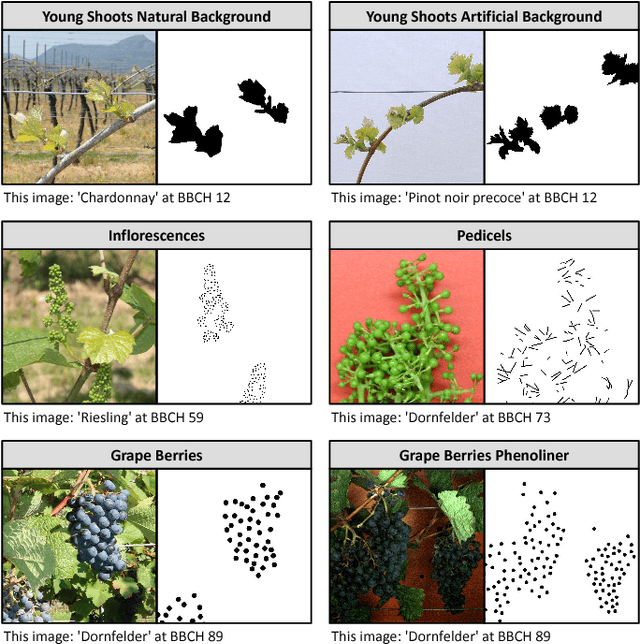
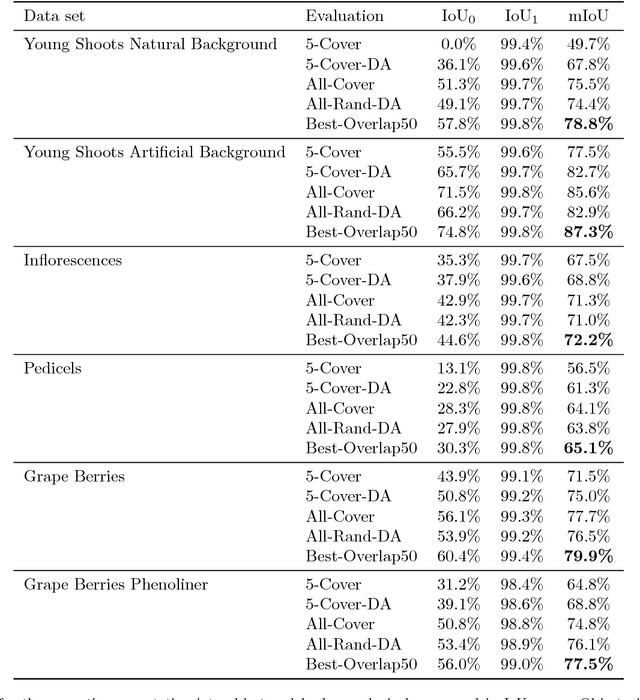
Abstract:In (grapevine) breeding programs and research, periodic phenotyping and multi-year monitoring of different grapevine traits, like growth or yield, is needed especially in the field. This demand imply objective, precise and automated methods using sensors and adaptive software. This work presents a proof-of-concept analyzing RGB images of different growth stages of grapevines with the aim to detect and quantify promising plant organs which are related to yield. The input images are segmented by a Fully Convolutional Neural Network (FCN) into object and background pixels. The objects are plant organs like young shoots, pedicels, flower buds or grapes, which are principally suitable for yield estimation. In the ground truth of the training images, each object is separately annotated as a connected segment of object pixels, which enables end-to-end learning of the object features. Based on the CNN-based segmentation, the number of objects is determined by detecting and counting connected components of object pixels using region labeling. In an evaluation on six different data sets, the system achieves an IoU of up to 87.3% for the segmentation and an F1 score of up to 88.6% for the object detection. The reason for the good results is the combination of a powerful CNN architecture and very precise and accurate image annotations.
Automated Phenotyping of Epicuticular Waxes of Grapevine Berries Using Light Separation and Convolutional Neural Networks
Sep 06, 2018



Abstract:In viticulture the epicuticular wax as the outer layer of the berry skin is known as trait which is correlated to resilience towards Botrytis bunch rot. Traditionally this trait is classified using the OIV descriptor 227 (berry bloom) in a time consuming way resulting in subjective and error-prone phenotypic data. In the present study an objective, fast and sensor-based approach was developed to monitor berry bloom. From the technical point-of-view, it is known that the measurement of different illumination components conveys important information about observed object surfaces. A Mobile Light-Separation-Lab is proposed in order to capture illumination-separated images of grapevine berries for phenotyping the distribution of epicuticular waxes (berry bloom). For image analysis, an efficient convolutional neural network approach is used to derive the uniformity and intactness of waxes on berries. Method validation over six grapevine cultivars shows accuracies up to $97.3$%. In addition, electrical impedance of the cuticle and its epicuticular waxes (described as an indicator for the thickness of berry skin and its permeability) was correlated to the detected proportion of waxes with $r=0.76$. This novel, fast and non-invasive phenotyping approach facilitates enlarged screenings within grapevine breeding material and genetic repositories regarding berry bloom characteristics and its impact on resilience towards Botrytis bunch rot.
Efficient identification, localization and quantification of grapevine inflorescences in unprepared field images using Fully Convolutional Networks
Jul 10, 2018



Abstract:Yield and its prediction is one of the most important tasks in grapevine breeding purposes and vineyard management. Commonly, this trait is estimated manually right before harvest by extrapolation, which mostly is labor-intensive, destructive and inaccurate. In the present study an automated image-based workflow was developed quantifying inflorescences and single flowers in unprepared field images of grapevines, i.e. no artificial background or light was applied. It is a novel approach for non-invasive, inexpensive and objective phenotyping with high-throughput. First, image regions depicting inflorescences were identified and localized. This was done by segmenting the images into the classes "inflorescence" and "non-inflorescence" using a Fully Convolutional Network (FCN). Efficient image segmentation hereby is the most challenging step regarding the small geometry and dense distribution of flowers (several hundred flowers per inflorescence), similar color of all plant organs in the fore- and background as well as the circumstance that only approximately 5% of an image show inflorescences. The trained FCN achieved a mean Intersection Over Union (IOU) of 87.6% on the test data set. Finally, individual flowers were extracted from the "inflorescence"-areas using Circular Hough Transform. The flower extraction achieved a recall of 80.3% and a precision of 70.7% using the segmentation derived by the trained FCN model. Summarized, the presented approach is a promising strategy in order to predict yield potential automatically in the earliest stage of grapevine development which is applicable for objective monitoring and evaluations of breeding material, genetic repositories or commercial vineyards.
Multi-View Semantic Labeling of 3D Point Clouds for Automated Plant Phenotyping
May 29, 2018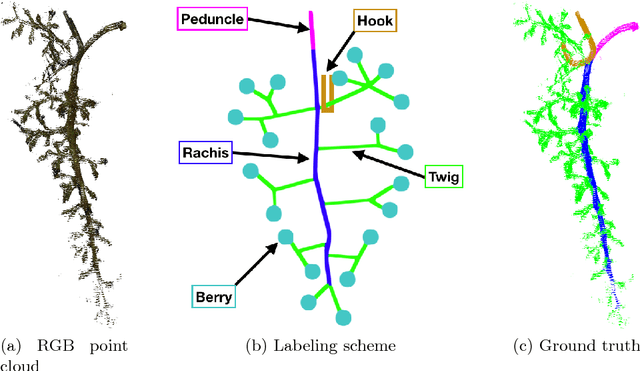
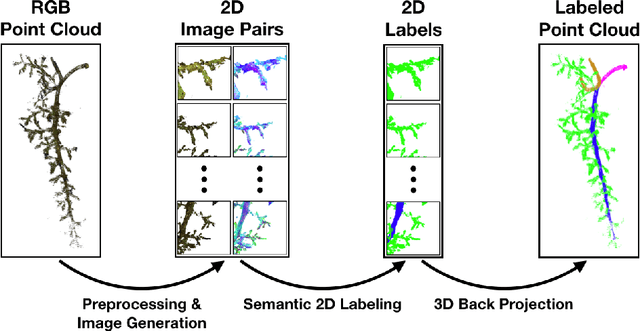

Abstract:Semantic labeling of 3D point clouds is important for the derivation of 3D models from real world scenarios in several economic fields such as building industry, facility management, town planning or heritage conservation. In contrast to these most common applications, we describe in this study the semantic labeling of 3D point clouds derived from plant organs by high-precision scanning. Our approach is optimized for the task of plant phenotyping with its very specific challenges and is employing a deep learning framework. Thereby, we report important experiences concerning detailed parameter initialization and optimization techniques. By evaluating our approach with challenging datasets we achieve state-of-the-art results without difficult and time consuming feature engineering as being necessary in traditional approaches to semantic labeling.
Automated Image Analysis Framework for the High-Throughput Determination of Grapevine Berry Sizes Using Conditional Random Fields
Dec 15, 2017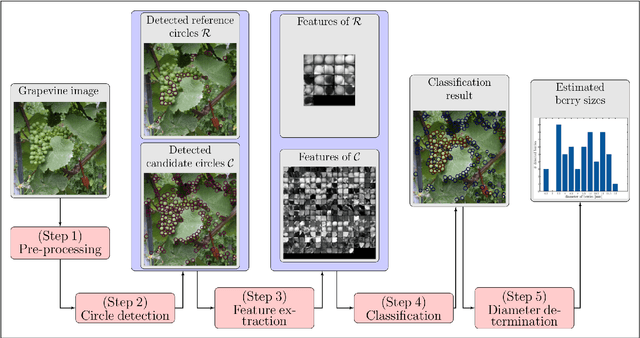
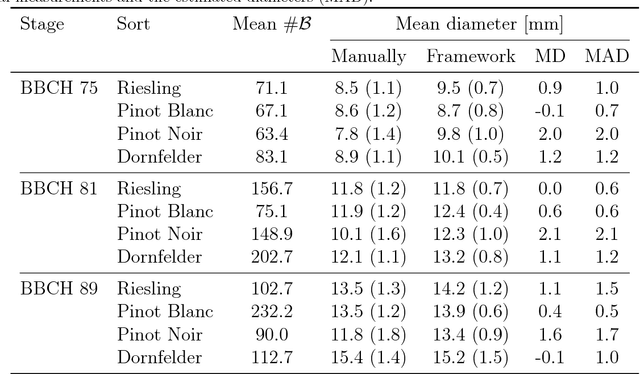
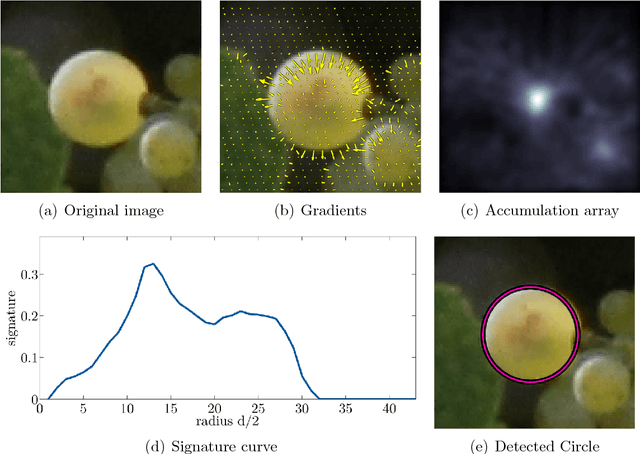
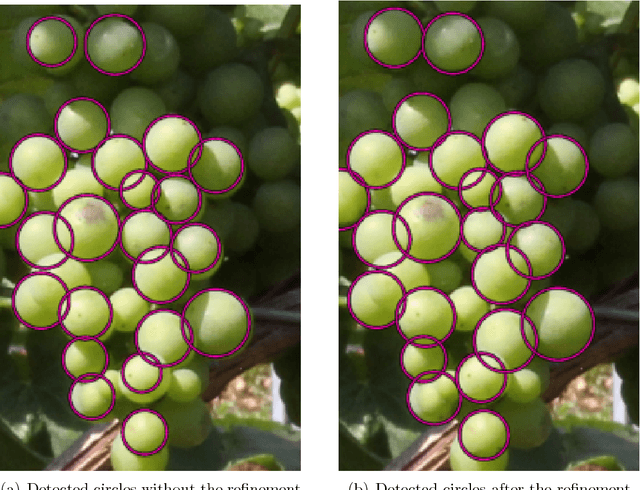
Abstract:The berry size is one of the most important fruit traits in grapevine breeding. Non-invasive, image-based phenotyping promises a fast and precise method for the monitoring of the grapevine berry size. In the present study an automated image analyzing framework was developed in order to estimate the size of grapevine berries from images in a high-throughput manner. The framework includes (i) the detection of circular structures which are potentially berries and (ii) the classification of these into the class 'berry' or 'non-berry' by utilizing a conditional random field. The approach used the concept of a one-class classification, since only the target class 'berry' is of interest and needs to be modeled. Moreover, the classification was carried out by using an automated active learning approach, i.e no user interaction is required during the classification process and in addition, the process adapts automatically to changing image conditions, e.g. illumination or berry color. The framework was tested on three datasets consisting in total of 139 images. The images were taken in an experimental vineyard at different stages of grapevine growth according to the BBCH scale. The mean berry size of a plant estimated by the framework correlates with the manually measured berry size by $0.88$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge