Karthick Gunasekaran
Diverse Distributions of Self-Supervised Tasks for Meta-Learning in NLP
Nov 02, 2021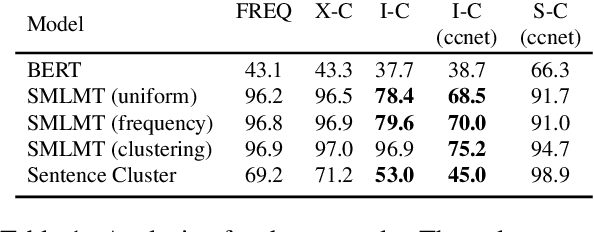
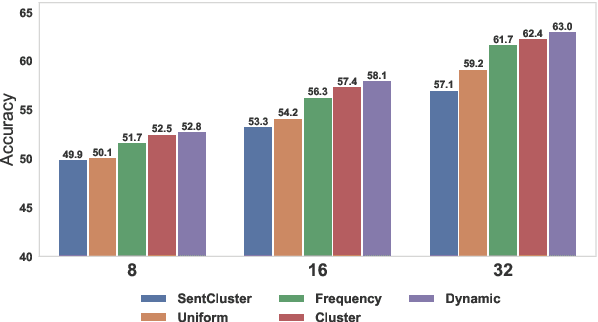
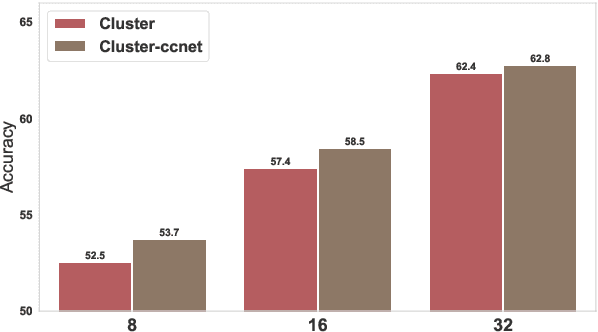
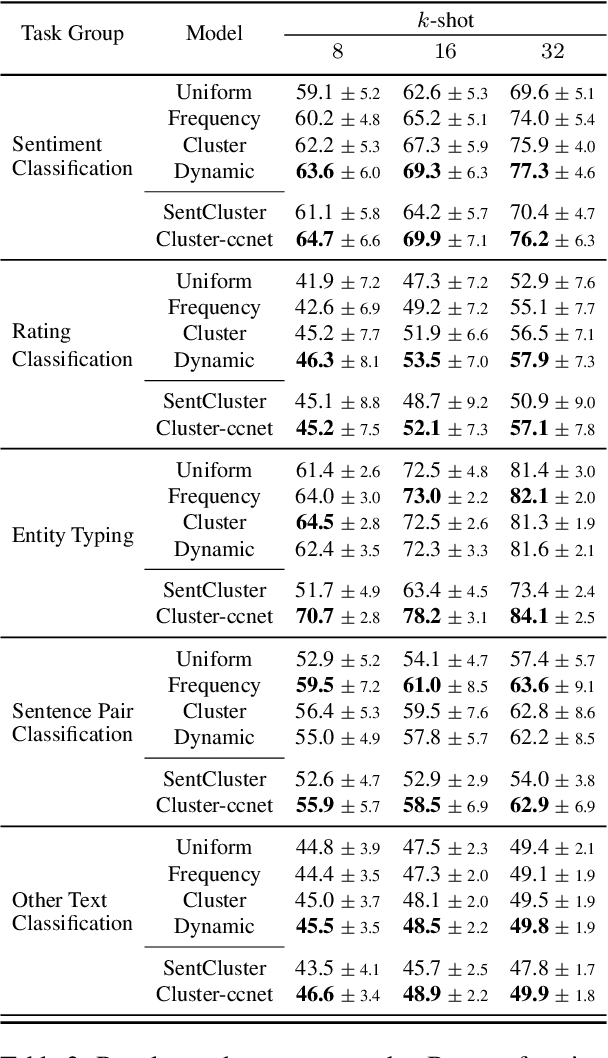
Abstract:Meta-learning considers the problem of learning an efficient learning process that can leverage its past experience to accurately solve new tasks. However, the efficacy of meta-learning crucially depends on the distribution of tasks available for training, and this is often assumed to be known a priori or constructed from limited supervised datasets. In this work, we aim to provide task distributions for meta-learning by considering self-supervised tasks automatically proposed from unlabeled text, to enable large-scale meta-learning in NLP. We design multiple distributions of self-supervised tasks by considering important aspects of task diversity, difficulty, type, domain, and curriculum, and investigate how they affect meta-learning performance. Our analysis shows that all these factors meaningfully alter the task distribution, some inducing significant improvements in downstream few-shot accuracy of the meta-learned models. Empirically, results on 20 downstream tasks show significant improvements in few-shot learning -- adding up to +4.2% absolute accuracy (on average) to the previous unsupervised meta-learning method, and perform comparably to supervised methods on the FewRel 2.0 benchmark.
Unsupervised Pre-training for Biomedical Question Answering
Sep 27, 2020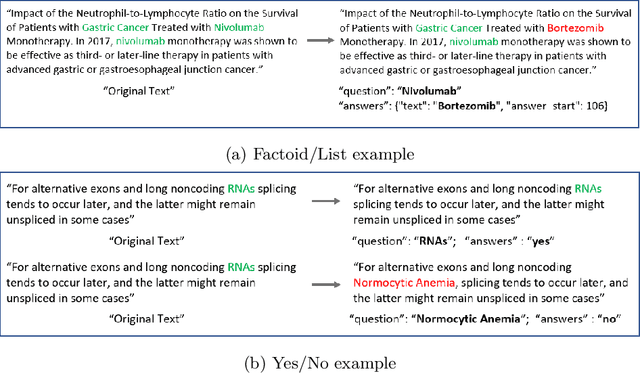
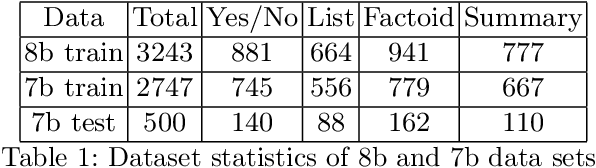
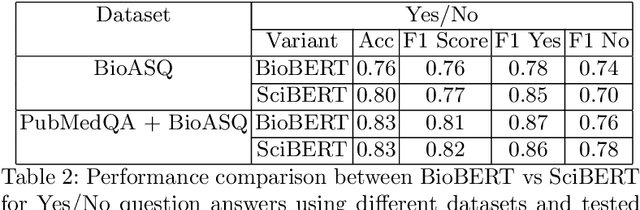
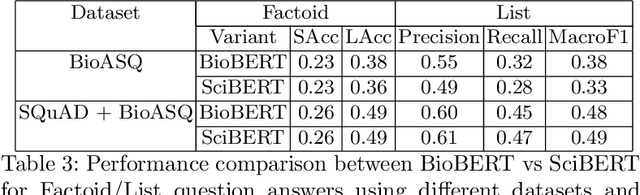
Abstract:We explore the suitability of unsupervised representation learning methods on biomedical text -- BioBERT, SciBERT, and BioSentVec -- for biomedical question answering. To further improve unsupervised representations for biomedical QA, we introduce a new pre-training task from unlabeled data designed to reason about biomedical entities in the context. Our pre-training method consists of corrupting a given context by randomly replacing some mention of a biomedical entity with a random entity mention and then querying the model with the correct entity mention in order to locate the corrupted part of the context. This de-noising task enables the model to learn good representations from abundant, unlabeled biomedical text that helps QA tasks and minimizes the train-test mismatch between the pre-training task and the downstream QA tasks by requiring the model to predict spans. Our experiments show that pre-training BioBERT on the proposed pre-training task significantly boosts performance and outperforms the previous best model from the 7th BioASQ Task 7b-Phase B challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge