Vaishnavi Kommaraju
k-Parameter Approach for False In-Season Anomaly Suppression in Daily Time Series Anomaly Detection
Nov 10, 2023



Abstract:Detecting anomalies in a daily time series with a weekly pattern is a common task with a wide range of applications. A typical way of performing the task is by using decomposition method. However, the method often generates false positive results where a data point falls within its weekly range but is just off from its weekday position. We refer to this type of anomalies as "in-season anomalies", and propose a k-parameter approach to address the issue. The approach provides configurable extra tolerance for in-season anomalies to suppress misleading alerts while preserving real positives. It yields favorable result.
Unsupervised Pre-training for Biomedical Question Answering
Sep 27, 2020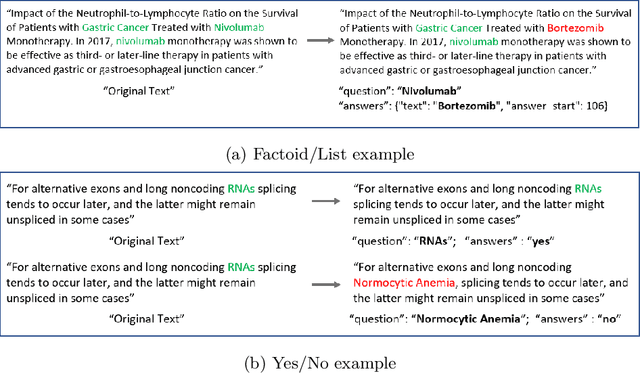
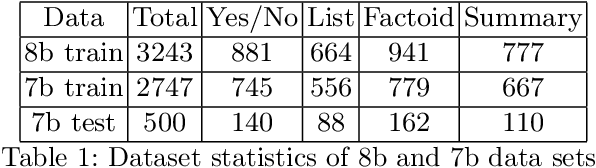
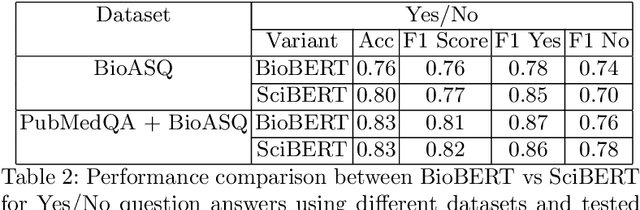
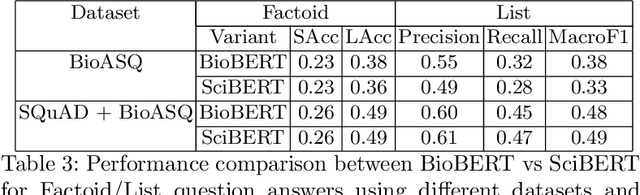
Abstract:We explore the suitability of unsupervised representation learning methods on biomedical text -- BioBERT, SciBERT, and BioSentVec -- for biomedical question answering. To further improve unsupervised representations for biomedical QA, we introduce a new pre-training task from unlabeled data designed to reason about biomedical entities in the context. Our pre-training method consists of corrupting a given context by randomly replacing some mention of a biomedical entity with a random entity mention and then querying the model with the correct entity mention in order to locate the corrupted part of the context. This de-noising task enables the model to learn good representations from abundant, unlabeled biomedical text that helps QA tasks and minimizes the train-test mismatch between the pre-training task and the downstream QA tasks by requiring the model to predict spans. Our experiments show that pre-training BioBERT on the proposed pre-training task significantly boosts performance and outperforms the previous best model from the 7th BioASQ Task 7b-Phase B challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge