Karoline Heiwolt
Lincoln's Annotated Spatio-Temporal Strawberry Dataset (LAST-Straw)
Mar 01, 2024Abstract:Automated phenotyping of plants for breeding and plant studies promises to provide quantitative metrics on plant traits at a previously unattainable observation frequency. Developers of tools for performing high-throughput phenotyping are, however, constrained by the availability of relevant datasets on which to perform validation. To this end, we present a spatio-temporal dataset of 3D point clouds of strawberry plants for two varieties, totalling 84 individual point clouds. We focus on the end use of such tools - the extraction of biologically relevant phenotypes - and demonstrate a phenotyping pipeline on the dataset. This comprises of the steps, including; segmentation, skeletonisation and tracking, and we detail how each stage facilitates the extraction of different phenotypes or provision of data insights. We particularly note that assessment is focused on the validation of phenotypes, extracted from the representations acquired at each step of the pipeline, rather than singularly focusing on assessing the representation itself. Therefore, where possible, we provide \textit{in silico} ground truth baselines for the phenotypes extracted at each step and introduce methodology for the quantitative assessment of skeletonisation and the length trait extracted thereof. This dataset contributes to the corpus of freely available agricultural/horticultural spatio-temporal data for the development of next-generation phenotyping tools, increasing the number of plant varieties available for research in this field and providing a basis for genuine comparison of new phenotyping methodology.
Statistical shape representations for temporal registration of plant components in 3D
Sep 23, 2022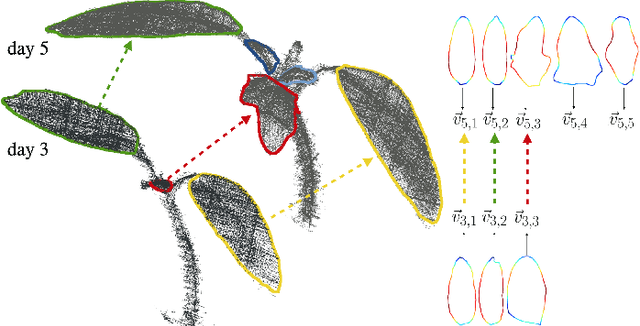
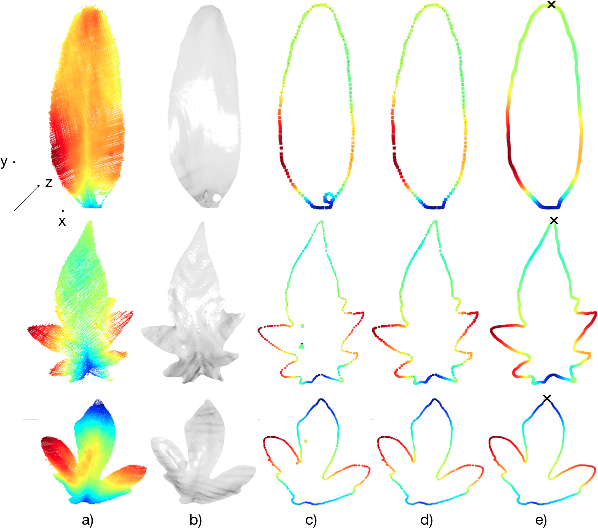
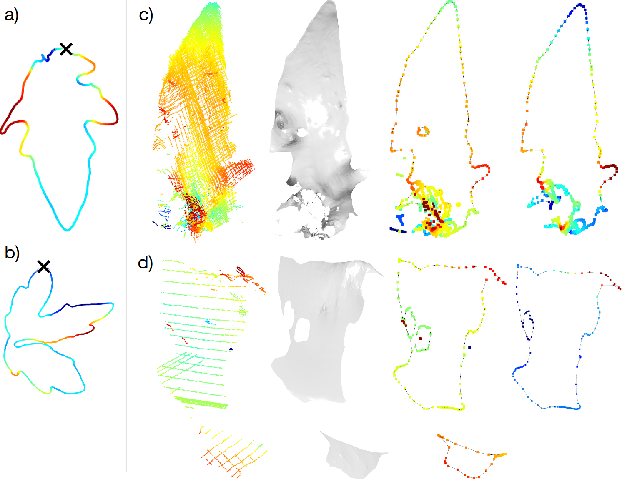
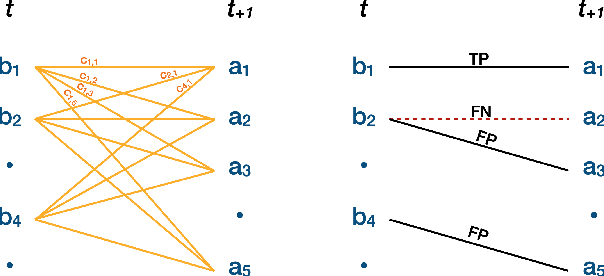
Abstract:Plants are dynamic organisms. Understanding temporal variations in vegetation is an essential problem for all robots in the wild. However, associating repeated 3D scans of plants across time is challenging. A key step in this process is re-identifying and tracking the same individual plant components over time. Previously, this has been achieved by comparing their global spatial or topological location. In this work, we demonstrate how using shape features improves temporal organ matching. We present a landmark-free shape compression algorithm, which allows for the extraction of 3D shape features of leaves, characterises leaf shape and curvature efficiently in few parameters, and makes the association of individual leaves in feature space possible. The approach combines 3D contour extraction and further compression using Principal Component Analysis (PCA) to produce a shape space encoding, which is entirely learned from data and retains information about edge contours and 3D curvature. Our evaluation on temporal scan sequences of tomato plants shows, that incorporating shape features improves temporal leaf-matching. A combination of shape, location, and rotation information proves most informative for recognition of leaves over time and yields a true positive rate of 75%, a 15% improvement on sate-of-the-art methods. This is essential for robotic crop monitoring, which enables whole-of-lifecycle phenotyping.
Automatic Detection of Myocontrol Failures Based upon Situational Context Information
Jun 27, 2019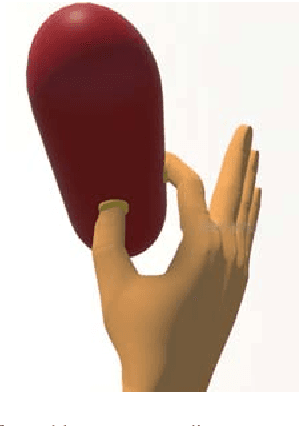
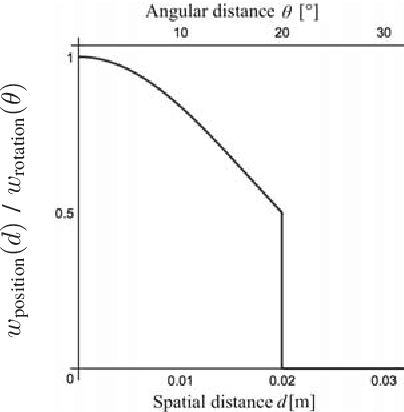
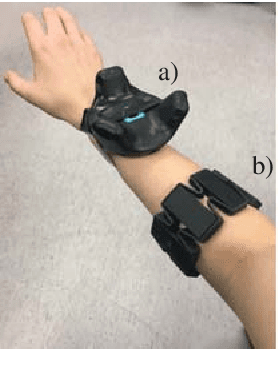
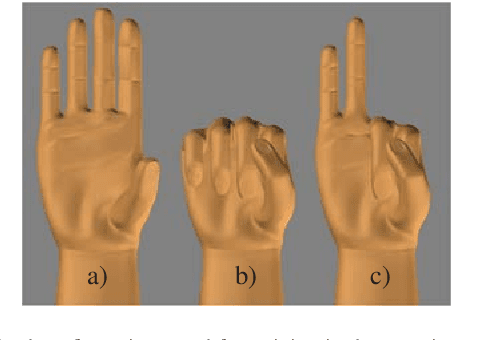
Abstract:Myoelectric control systems for assistive devices are still unreliable. The user's input signals can become unstable over time due to e.g. fatigue, electrode displacement, or sweat. Hence, such controllers need to be constantly updated and heavily rely on user feedback. In this paper, we present an automatic failure detection method which learns when plausible predictions become unreliable and model updates are necessary. Our key insight is to enhance the control system with a set of generative models that learn sensible behaviour for a desired task from human demonstration. We illustrate our approach on a grasping scenario in Virtual Reality, in which the user is asked to grasp a bottle on a table. From demonstration our model learns the reach-to-grasp motion from a resting position to two grasps (power grasp and tridigital grasp) and how to predict the most adequate grasp from local context, e.g. tridigital grasp on the bottle cap or around the bottleneck. By measuring the error between new grasp attempts and the model prediction, the system can effectively detect which input commands do not reflect the user's intention. We evaluated our model in two cases: i) with both position and rotation information of the wrist pose, and ii) with only rotational information. Our results show that our approach detects statistically highly significant differences in error distributions with p < 0.001 between successful and failed grasp attempts in both cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge