Karen O'Connor
Reddit-Impacts: A Named Entity Recognition Dataset for Analyzing Clinical and Social Effects of Substance Use Derived from Social Media
May 09, 2024



Abstract:Substance use disorders (SUDs) are a growing concern globally, necessitating enhanced understanding of the problem and its trends through data-driven research. Social media are unique and important sources of information about SUDs, particularly since the data in such sources are often generated by people with lived experiences. In this paper, we introduce Reddit-Impacts, a challenging Named Entity Recognition (NER) dataset curated from subreddits dedicated to discussions on prescription and illicit opioids, as well as medications for opioid use disorder. The dataset specifically concentrates on the lesser-studied, yet critically important, aspects of substance use--its clinical and social impacts. We collected data from chosen subreddits using the publicly available Application Programming Interface for Reddit. We manually annotated text spans representing clinical and social impacts reported by people who also reported personal nonmedical use of substances including but not limited to opioids, stimulants and benzodiazepines. Our objective is to create a resource that can enable the development of systems that can automatically detect clinical and social impacts of substance use from text-based social media data. The successful development of such systems may enable us to better understand how nonmedical use of substances affects individual health and societal dynamics, aiding the development of effective public health strategies. In addition to creating the annotated data set, we applied several machine learning models to establish baseline performances. Specifically, we experimented with transformer models like BERT, and RoBERTa, one few-shot learning model DANN by leveraging the full training dataset, and GPT-3.5 by using one-shot learning, for automatic NER of clinical and social impacts. The dataset has been made available through the 2024 SMM4H shared tasks.
Deep Neural Networks Ensemble for Detecting Medication Mentions in Tweets
Apr 10, 2019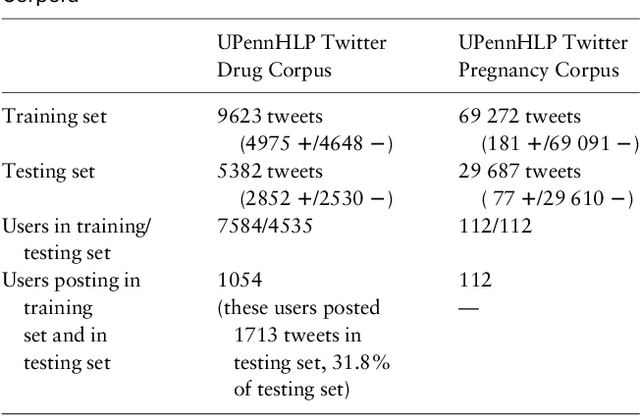


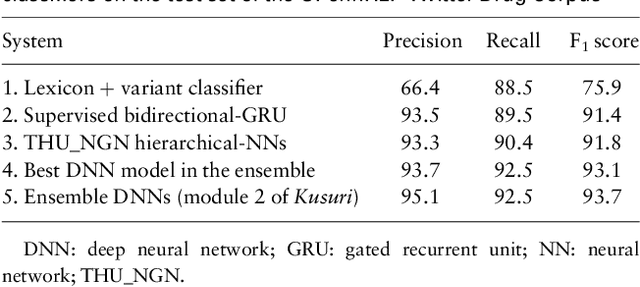
Abstract:Objective: After years of research, Twitter posts are now recognized as an important source of patient-generated data, providing unique insights into population health. A fundamental step to incorporating Twitter data in pharmacoepidemiological research is to automatically recognize medication mentions in tweets. Given that lexical searches for medication names may fail due to misspellings or ambiguity with common words, we propose a more advanced method to recognize them. Methods: We present Kusuri, an Ensemble Learning classifier, able to identify tweets mentioning drug products and dietary supplements. Kusuri ("medication" in Japanese) is composed of two modules. First, four different classifiers (lexicon-based, spelling-variant-based, pattern-based and one based on a weakly-trained neural network) are applied in parallel to discover tweets potentially containing medication names. Second, an ensemble of deep neural networks encoding morphological, semantical and long-range dependencies of important words in the tweets discovered is used to make the final decision. Results: On a balanced (50-50) corpus of 15,005 tweets, Kusuri demonstrated performances close to human annotators with 93.7% F1-score, the best score achieved thus far on this corpus. On a corpus made of all tweets posted by 113 Twitter users (98,959 tweets, with only 0.26% mentioning medications), Kusuri obtained 76.3% F1-score. There is not a prior drug extraction system that compares running on such an extremely unbalanced dataset. Conclusion: The system identifies tweets mentioning drug names with performance high enough to ensure its usefulness and ready to be integrated in larger natural language processing systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge