June-Goo Lee
CycleMorph: Cycle Consistent Unsupervised Deformable Image Registration
Aug 13, 2020
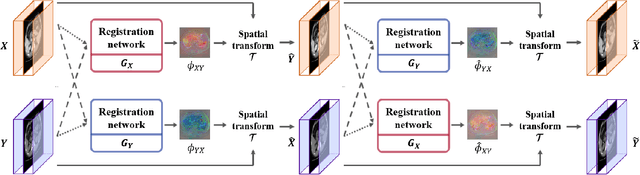
Abstract:Image registration is a fundamental task in medical image analysis. Recently, deep learning based image registration methods have been extensively investigated due to their excellent performance despite the ultra-fast computational time. However, the existing deep learning methods still have limitation in the preservation of original topology during the deformation with registration vector fields. To address this issues, here we present a cycle-consistent deformable image registration. The cycle consistency enhances image registration performance by providing an implicit regularization to preserve topology during the deformation. The proposed method is so flexible that can be applied for both 2D and 3D registration problems for various applications, and can be easily extended to multi-scale implementation to deal with the memory issues in large volume registration. Experimental results on various datasets from medical and non-medical applications demonstrate that the proposed method provides effective and accurate registration on diverse image pairs within a few seconds. Qualitative and quantitative evaluations on deformation fields also verify the effectiveness of the cycle consistency of the proposed method.
Unsupervised Deformable Image Registration Using Cycle-Consistent CNN
Jul 02, 2019
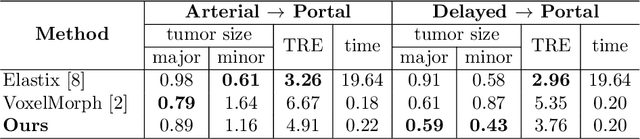
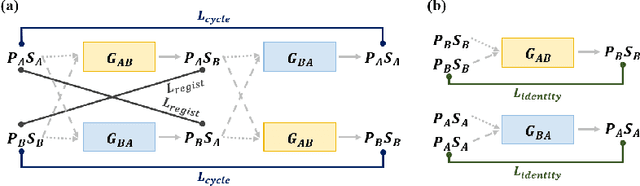
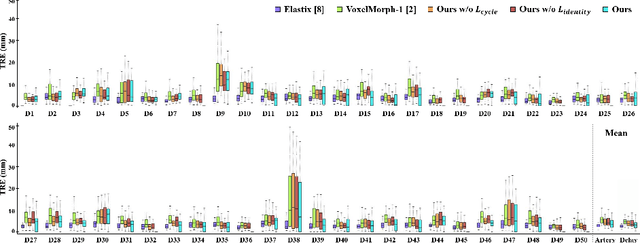
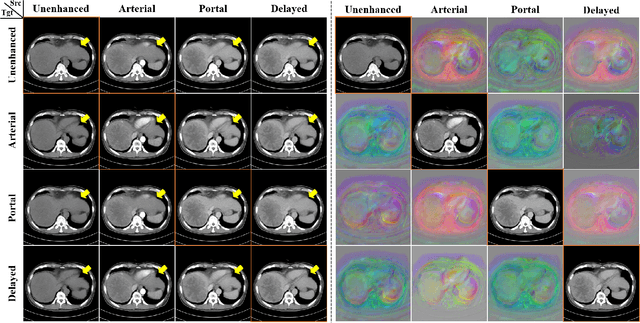
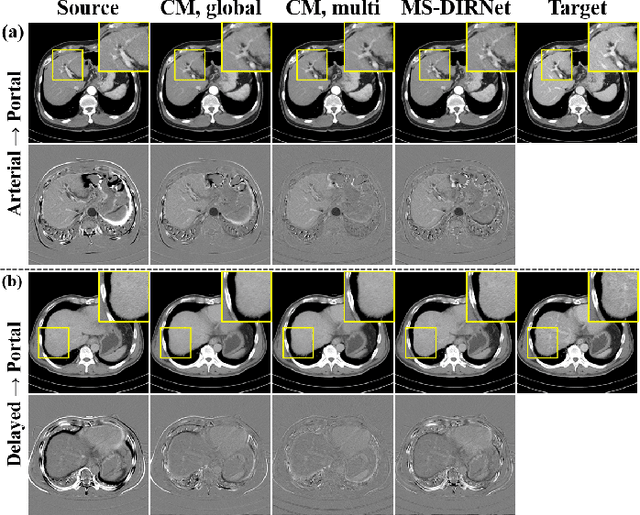
Abstract:Medical image registration is one of the key processing steps for biomedical image analysis such as cancer diagnosis. Recently, deep learning based supervised and unsupervised image registration methods have been extensively studied due to its excellent performance in spite of ultra-fast computational time compared to the classical approaches. In this paper, we present a novel unsupervised medical image registration method that trains deep neural network for deformable registration of 3D volumes using a cycle-consistency. Thanks to the cycle consistency, the proposed deep neural networks can take diverse pair of image data with severe deformation for accurate registration. Experimental results using multiphase liver CT images demonstrate that our method provides very precise 3D image registration within a few seconds, resulting in more accurate cancer size estimation.
Automated detection of vulnerable plaque in intravascular ultrasound images
Apr 18, 2018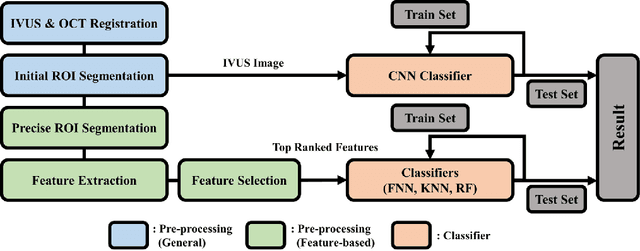
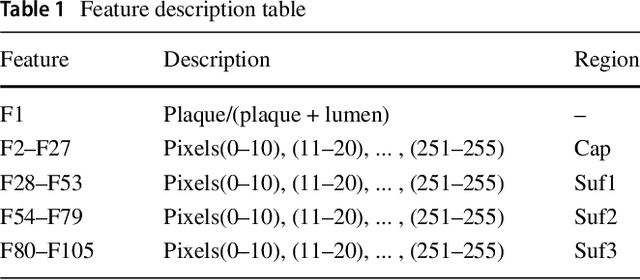
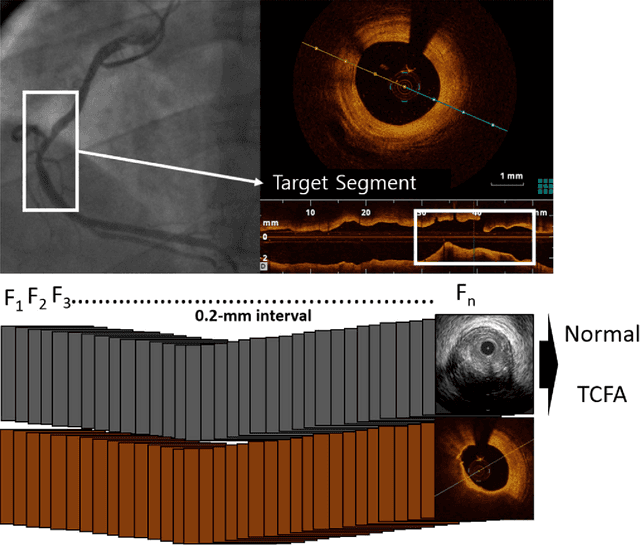
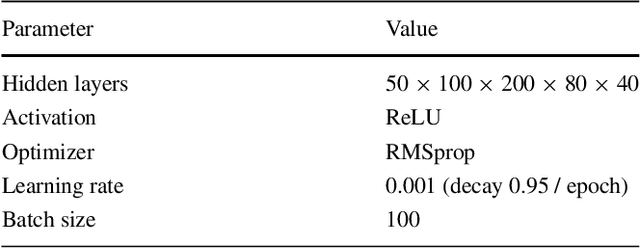
Abstract:Acute Coronary Syndrome (ACS) is a syndrome caused by a decrease in blood flow in the coronary arteries. The ACS is usually related to coronary thrombosis and is primarily caused by plaque rupture followed by plaque erosion and calcified nodule. Thin-cap fibroatheroma (TCFA) is known to be the most similar lesion morphologically to a plaque rupture. In this paper, we propose methods to classify TCFA using various machine learning classifiers including Feed-forward Neural Network (FNN), K-Nearest Neighbor (KNN), Random Forest (RF) and Convolutional Neural Network (CNN) to figure out a classifier that shows optimal TCFA classification accuracy. In addition, we suggest pixel range based feature extraction method to extract the ratio of pixels in the different region of interests to reflect the physician's TCFA discrimination criteria. A total of 12,325 IVUS images were labeled with corresponding OCT images to train and evaluate the classifiers. We achieved 0.884, 0.890, 0.878 and 0.933 Area Under the ROC Curve (AUC) in the order of using FNN, KNN, RF and CNN classifier. As a result, the CNN classifier performed best and the top 10 features of the feature-based classifiers (FNN, KNN, RF) were found to be similar to the physician's TCFA diagnostic criteria.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge