Joshua Wong
A Reproducible Framework for Bias-Resistant Machine Learning on Small-Sample Neuroimaging Data
Feb 02, 2026Abstract:We introduce a reproducible, bias-resistant machine learning framework that integrates domain-informed feature engineering, nested cross-validation, and calibrated decision-threshold optimization for small-sample neuroimaging data. Conventional cross-validation frameworks that reuse the same folds for both model selection and performance estimation yield optimistically biased results, limiting reproducibility and generalization. Demonstrated on a high-dimensional structural MRI dataset of deep brain stimulation cognitive outcomes, the framework achieved a nested-CV balanced accuracy of 0.660\,$\pm$\,0.068 using a compact, interpretable subset selected via importance-guided ranking. By combining interpretability and unbiased evaluation, this work provides a generalizable computational blueprint for reliable machine learning in data-limited biomedical domains.
A Multi-scale Linear-time Encoder for Whole-Slide Image Analysis
Feb 02, 2026Abstract:We introduce Multi-scale Adaptive Recurrent Biomedical Linear-time Encoder (MARBLE), the first \textit{purely Mamba-based} multi-state multiple instance learning (MIL) framework for whole-slide image (WSI) analysis. MARBLE processes multiple magnification levels in parallel and integrates coarse-to-fine reasoning within a linear-time state-space model, efficiently capturing cross-scale dependencies with minimal parameter overhead. WSI analysis remains challenging due to gigapixel resolutions and hierarchical magnifications, while existing MIL methods typically operate at a single scale and transformer-based approaches suffer from quadratic attention costs. By coupling parallel multi-scale processing with linear-time sequence modeling, MARBLE provides a scalable and modular alternative to attention-based architectures. Experiments on five public datasets show improvements of up to \textbf{6.9\%} in AUC, \textbf{20.3\%} in accuracy, and \textbf{2.3\%} in C-index, establishing MARBLE as an efficient and generalizable framework for multi-scale WSI analysis.
Relation Extraction from Tables using Artificially Generated Metadata
Sep 06, 2021



Abstract:Relation Extraction (RE) from tables is the task of identifying relations between pairs of columns of a table. Generally, RE models for this task require labelled tables for training. These labelled tables can also be generated artificially from a Knowledge Graph (KG), which makes the cost to acquire them much lower in comparison to manual annotations. However, unlike real tables, these synthetic tables lack associated metadata, such as, column-headers, captions, etc; this is because synthetic tables are created out of KGs that do not store such metadata. Meanwhile, previous works have shown that metadata is important for accurate RE from tables. To address this issue, we propose methods to artificially create some of this metadata for synthetic tables. Afterward, we experiment with a BERT-based model, in line with recently published works, that takes as input a combination of proposed artificial metadata and table content. Our empirical results show that this leads to an improvement of 9\%-45\% in F1 score, in absolute terms, over 2 tabular datasets.
Chemical Structure Elucidation from Mass Spectrometry by Matching Substructures
Nov 17, 2018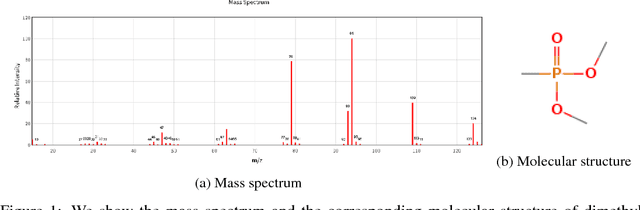
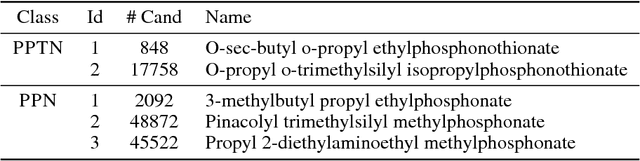
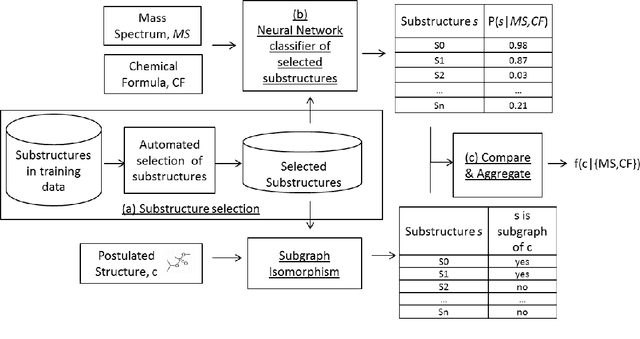
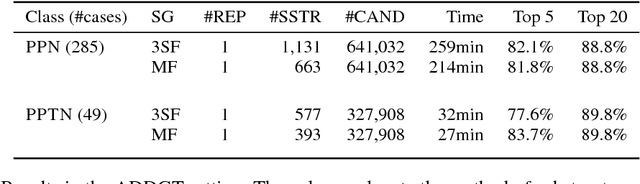
Abstract:Chemical structure elucidation is a serious bottleneck in analytical chemistry today. We address the problem of identifying an unknown chemical threat given its mass spectrum and its chemical formula, a task which might take well trained chemists several days to complete. Given a chemical formula, there could be over a million possible candidate structures. We take a data driven approach to rank these structures by using neural networks to predict the presence of substructures given the mass spectrum, and matching these substructures to the candidate structures. Empirically, we evaluate our approach on a data set of chemical agents built for unknown chemical threat identification. We show that our substructure classifiers can attain over 90% micro F1-score, and we can find the correct structure among the top 20 candidates in 88% and 71% of test cases for two compound classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge