Jonathan Henrich
Benchmarking pig detection and tracking under diverse and challenging conditions
Jul 22, 2025Abstract:To ensure animal welfare and effective management in pig farming, monitoring individual behavior is a crucial prerequisite. While monitoring tasks have traditionally been carried out manually, advances in machine learning have made it possible to collect individualized information in an increasingly automated way. Central to these methods is the localization of animals across space (object detection) and time (multi-object tracking). Despite extensive research of these two tasks in pig farming, a systematic benchmarking study has not yet been conducted. In this work, we address this gap by curating two datasets: PigDetect for object detection and PigTrack for multi-object tracking. The datasets are based on diverse image and video material from realistic barn conditions, and include challenging scenarios such as occlusions or bad visibility. For object detection, we show that challenging training images improve detection performance beyond what is achievable with randomly sampled images alone. Comparing different approaches, we found that state-of-the-art models offer substantial improvements in detection quality over real-time alternatives. For multi-object tracking, we observed that SORT-based methods achieve superior detection performance compared to end-to-end trainable models. However, end-to-end models show better association performance, suggesting they could become strong alternatives in the future. We also investigate characteristic failure cases of end-to-end models, providing guidance for future improvements. The detection and tracking models trained on our datasets perform well in unseen pens, suggesting good generalization capabilities. This highlights the importance of high-quality training data. The datasets and research code are made publicly available to facilitate reproducibility, re-use and further development.
Towards general deep-learning-based tree instance segmentation models
May 03, 2024Abstract:The segmentation of individual trees from forest point clouds is a crucial task for downstream analyses such as carbon sequestration estimation. Recently, deep-learning-based methods have been proposed which show the potential of learning to segment trees. Since these methods are trained in a supervised way, the question arises how general models can be obtained that are applicable across a wide range of settings. So far, training has been mainly conducted with data from one specific laser scanning type and for specific types of forests. In this work, we train one segmentation model under various conditions, using seven diverse datasets found in literature, to gain insights into the generalization capabilities under domain-shift. Our results suggest that a generalization from coniferous dominated sparse point clouds to deciduous dominated high-resolution point clouds is possible. Conversely, qualitative evidence suggests that generalization from high-resolution to low-resolution point clouds is challenging. This emphasizes the need for forest point clouds with diverse data characteristics for model development. To enrich the available data basis, labeled trees from two previous works were propagated to the complete forest point cloud and are made publicly available at https://doi.org/10.25625/QUTUWU.
Computer Vision for Primate Behavior Analysis in the Wild
Jan 29, 2024Abstract:Advances in computer vision as well as increasingly widespread video-based behavioral monitoring have great potential for transforming how we study animal cognition and behavior. However, there is still a fairly large gap between the exciting prospects and what can actually be achieved in practice today, especially in videos from the wild. With this perspective paper, we want to contribute towards closing this gap, by guiding behavioral scientists in what can be expected from current methods and steering computer vision researchers towards problems that are relevant to advance research in animal behavior. We start with a survey of the state-of-the-art methods for computer vision problems that are directly relevant to the video-based study of animal behavior, including object detection, multi-individual tracking, (inter)action recognition and individual identification. We then review methods for effort-efficient learning, which is one of the biggest challenges from a practical perspective. Finally, we close with an outlook into the future of the emerging field of computer vision for animal behavior, where we argue that the field should move fast beyond the common frame-by-frame processing and treat video as a first-class citizen.
TreeLearn: A Comprehensive Deep Learning Method for Segmenting Individual Trees from Forest Point Clouds
Sep 15, 2023

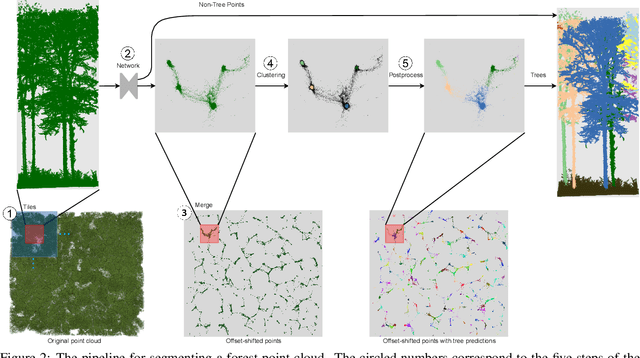

Abstract:Laser-scanned point clouds of forests make it possible to extract valuable information for forest management. To consider single trees, a forest point cloud needs to be segmented into individual tree point clouds. Existing segmentation methods are usually based on hand-crafted algorithms, such as identifying trunks and growing trees from them, and face difficulties in dense forests with overlapping tree crowns. In this study, we propose \mbox{TreeLearn}, a deep learning-based approach for semantic and instance segmentation of forest point clouds. Unlike previous methods, TreeLearn is trained on already segmented point clouds in a data-driven manner, making it less reliant on predefined features and algorithms. Additionally, we introduce a new manually segmented benchmark forest dataset containing 156 full trees, and 79 partial trees, that have been cleanly segmented by hand. This enables the evaluation of instance segmentation performance going beyond just evaluating the detection of individual trees. We trained TreeLearn on forest point clouds of 6665 trees, labeled using the Lidar360 software. An evaluation on the benchmark dataset shows that TreeLearn performs equally well or better than the algorithm used to generate its training data. Furthermore, the method's performance can be vastly improved by fine-tuning on the cleanly labeled benchmark dataset. The TreeLearn code is availabe from https://github.com/ecker-lab/TreeLearn. The data as well as trained models can be found at https://doi.org/10.25625/VPMPID.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge