John S. Brownstein
TCube: Domain-Agnostic Neural Time-series Narration
Oct 11, 2021



Abstract:The task of generating rich and fluent narratives that aptly describe the characteristics, trends, and anomalies of time-series data is invaluable to the sciences (geology, meteorology, epidemiology) or finance (trades, stocks, or sales and inventory). The efforts for time-series narration hitherto are domain-specific and use predefined templates that offer consistency but lead to mechanical narratives. We present TCube (Time-series-to-text), a domain-agnostic neural framework for time-series narration, that couples the representation of essential time-series elements in the form of a dense knowledge graph and the translation of said knowledge graph into rich and fluent narratives through the transfer-learning capabilities of PLMs (Pre-trained Language Models). TCube's design primarily addresses the challenge that lies in building a neural framework in the complete paucity of annotated training data for time-series. The design incorporates knowledge graphs as an intermediary for the representation of essential time-series elements which can be linearized for textual translation. To the best of our knowledge, TCube is the first investigation of the use of neural strategies for time-series narration. Through extensive evaluations, we show that TCube can improve the lexical diversity of the generated narratives by up to 65.38% while still maintaining grammatical integrity. The practicality and deployability of TCube is further validated through an expert review (n=21) where 76.2% of participating experts wary of auto-generated narratives favored TCube as a deployable system for time-series narration due to its richer narratives. Our code-base, models, and datasets, with detailed instructions for reproducibility is publicly hosted at https://github.com/Mandar-Sharma/TCube.
Guided Deep List: Automating the Generation of Epidemiological Line Lists from Open Sources
Feb 22, 2017
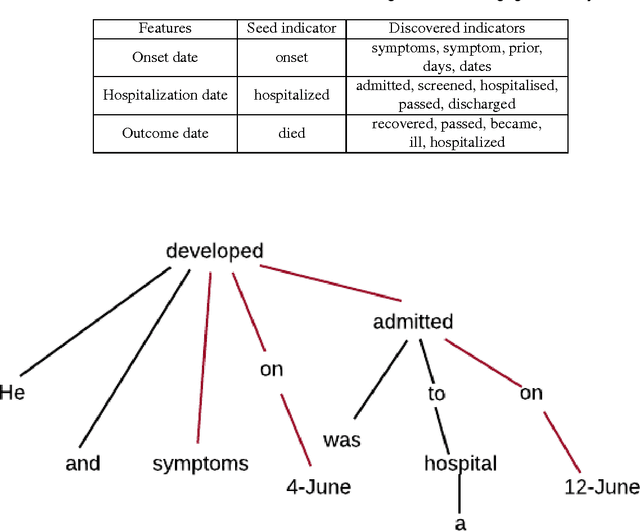

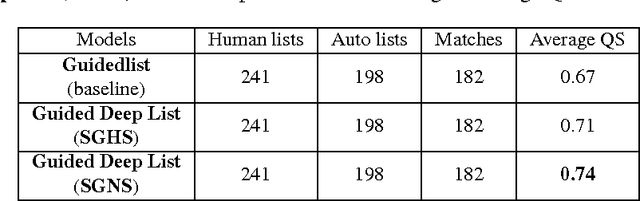
Abstract:Real-time monitoring and responses to emerging public health threats rely on the availability of timely surveillance data. During the early stages of an epidemic, the ready availability of line lists with detailed tabular information about laboratory-confirmed cases can assist epidemiologists in making reliable inferences and forecasts. Such inferences are crucial to understand the epidemiology of a specific disease early enough to stop or control the outbreak. However, construction of such line lists requires considerable human supervision and therefore, difficult to generate in real-time. In this paper, we motivate Guided Deep List, the first tool for building automated line lists (in near real-time) from open source reports of emerging disease outbreaks. Specifically, we focus on deriving epidemiological characteristics of an emerging disease and the affected population from reports of illness. Guided Deep List uses distributed vector representations (ala word2vec) to discover a set of indicators for each line list feature. This discovery of indicators is followed by the use of dependency parsing based techniques for final extraction in tabular form. We evaluate the performance of Guided Deep List against a human annotated line list provided by HealthMap corresponding to MERS outbreaks in Saudi Arabia. We demonstrate that Guided Deep List extracts line list features with increased accuracy compared to a baseline method. We further show how these automatically extracted line list features can be used for making epidemiological inferences, such as inferring demographics and symptoms-to-hospitalization period of affected individuals.
Characterizing Diseases from Unstructured Text: A Vocabulary Driven Word2vec Approach
Jun 03, 2016
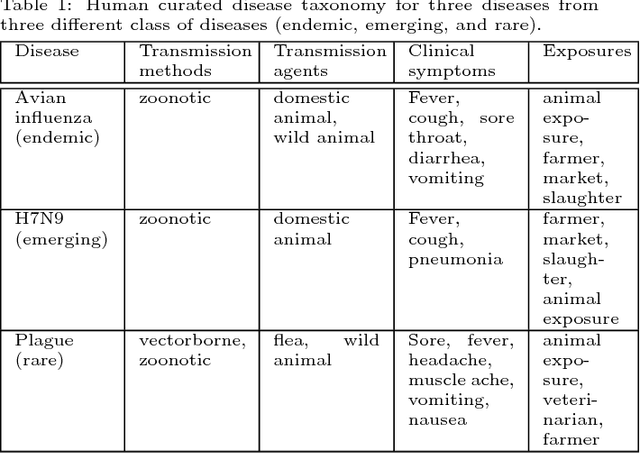

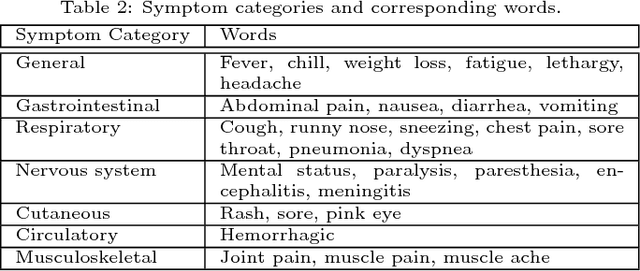
Abstract:Traditional disease surveillance can be augmented with a wide variety of real-time sources such as, news and social media. However, these sources are in general unstructured and, construction of surveillance tools such as taxonomical correlations and trace mapping involves considerable human supervision. In this paper, we motivate a disease vocabulary driven word2vec model (Dis2Vec) to model diseases and constituent attributes as word embeddings from the HealthMap news corpus. We use these word embeddings to automatically create disease taxonomies and evaluate our model against corresponding human annotated taxonomies. We compare our model accuracies against several state-of-the art word2vec methods. Our results demonstrate that Dis2Vec outperforms traditional distributed vector representations in its ability to faithfully capture taxonomical attributes across different class of diseases such as endemic, emerging and rare.
Temporal Topic Modeling to Assess Associations between News Trends and Infectious Disease Outbreaks
Jun 01, 2016



Abstract:In retrospective assessments, internet news reports have been shown to capture early reports of unknown infectious disease transmission prior to official laboratory confirmation. In general, media interest and reporting peaks and wanes during the course of an outbreak. In this study, we quantify the extent to which media interest during infectious disease outbreaks is indicative of trends of reported incidence. We introduce an approach that uses supervised temporal topic models to transform large corpora of news articles into temporal topic trends. The key advantages of this approach include, applicability to a wide range of diseases, and ability to capture disease dynamics - including seasonality, abrupt peaks and troughs. We evaluated the method using data from multiple infectious disease outbreaks reported in the United States of America (U.S.), China and India. We noted that temporal topic trends extracted from disease-related news reports successfully captured the dynamics of multiple outbreaks such as whooping cough in U.S. (2012), dengue outbreaks in India (2013) and China (2014). Our observations also suggest that efficient modeling of temporal topic trends using time-series regression techniques can estimate disease case counts with increased precision before official reports by health organizations.
Cloud-based Electronic Health Records for Real-time, Region-specific Influenza Surveillance
Dec 13, 2015



Abstract:Accurate real-time monitoring systems of influenza outbreaks help public health officials make informed decisions that may help save lives. We show that information extracted from cloud-based electronic health records databases, in combination with machine learning techniques and historical epidemiological information, have the potential to accurately and reliably provide near real-time regional predictions of flu outbreaks in the United States.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge