Characterizing Diseases from Unstructured Text: A Vocabulary Driven Word2vec Approach
Paper and Code
Jun 03, 2016
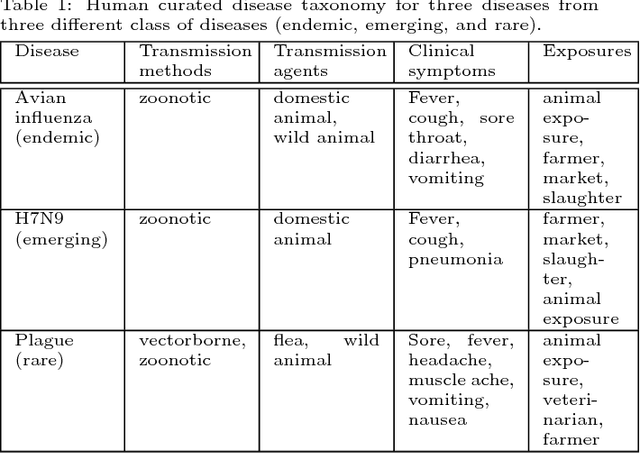

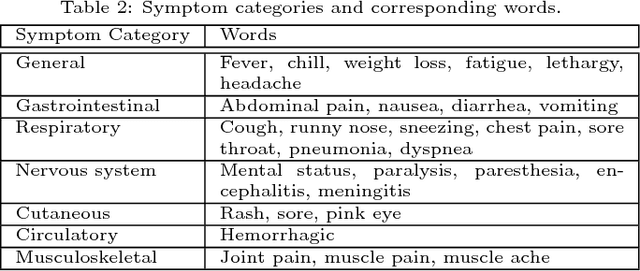
Traditional disease surveillance can be augmented with a wide variety of real-time sources such as, news and social media. However, these sources are in general unstructured and, construction of surveillance tools such as taxonomical correlations and trace mapping involves considerable human supervision. In this paper, we motivate a disease vocabulary driven word2vec model (Dis2Vec) to model diseases and constituent attributes as word embeddings from the HealthMap news corpus. We use these word embeddings to automatically create disease taxonomies and evaluate our model against corresponding human annotated taxonomies. We compare our model accuracies against several state-of-the art word2vec methods. Our results demonstrate that Dis2Vec outperforms traditional distributed vector representations in its ability to faithfully capture taxonomical attributes across different class of diseases such as endemic, emerging and rare.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge