Jochen B. Fiebach
Anonymization of labeled TOF-MRA images for brain vessel segmentation using generative adversarial networks
Sep 16, 2020



Abstract:Anonymization and data sharing are crucial for privacy protection and acquisition of large datasets for medical image analysis. This is a big challenge, especially for neuroimaging. Here, the brain's unique structure allows for re-identification and thus requires non-conventional anonymization. Generative adversarial networks (GANs) have the potential to provide anonymous images while preserving predictive properties. Analyzing brain vessel segmentation as a use case, we trained 3 GANs on time-of-flight (TOF) magnetic resonance angiography (MRA) patches for image-label generation: 1) Deep convolutional GAN, 2) Wasserstein-GAN with gradient penalty (WGAN-GP) and 3) WGAN-GP with spectral normalization (WGAN-GP-SN). The generated image-labels from each GAN were used to train a U-net for segmentation and tested on real data. Moreover, we applied our synthetic patches using transfer learning on a second dataset. For an increasing number of up to 15 patients we evaluated the model performance on real data with and without pre-training. The performance for all models was assessed by the Dice Similarity Coefficient (DSC) and the 95th percentile of the Hausdorff Distance (95HD). Comparing the 3 GANs, the U-net trained on synthetic data generated by the WGAN-GP-SN showed the highest performance to predict vessels (DSC/95HD 0.82/28.97) benchmarked by the U-net trained on real data (0.89/26.61). The transfer learning approach showed superior performance for the same GAN compared to no pre-training, especially for one patient only (0.91/25.68 vs. 0.85/27.36). In this work, synthetic image-label pairs retained generalizable information and showed good performance for vessel segmentation. Besides, we showed that synthetic patches can be used in a transfer learning approach with independent data. This paves the way to overcome the challenges of scarce data and anonymization in medical imaging.
On The Usage Of Average Hausdorff Distance For Segmentation Performance Assessment: Hidden Bias When Used For Ranking
Sep 13, 2020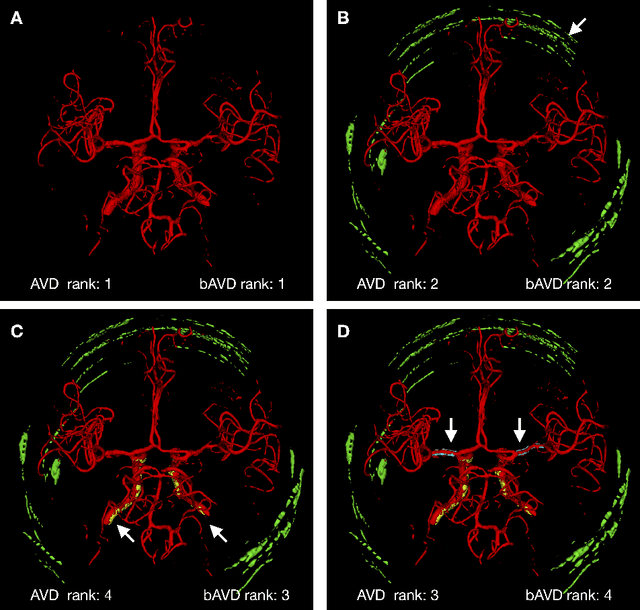
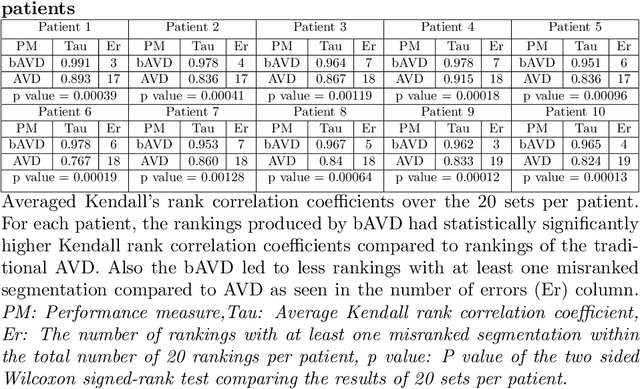
Abstract:Average Hausdorff Distance (AVD) is a widely used performance measure to calculate the distance between two point sets. In medical image segmentation, AVD is used to compare ground truth images with segmentation results allowing their ranking. We identified, however, a ranking bias of AVD making it less suitable for segmentation ranking. To mitigate this bias, we present a modified calculation of AVD that we have coined balanced AVD (bAVD). To simulate segmentations for ranking, we manually created non-overlapping segmentation errors common in cerebral vessel segmentation as our use-case. Adding the created errors consecutively and randomly to the ground truth, we created sets of simulated segmentations with increasing number of errors. Each set of simulated segmentations was ranked using AVD and bAVD. We calculated the Kendall-rank-correlation-coefficient between the segmentation ranking and the number of errors in each simulated segmentation. The rankings produced by bAVD had a significantly higher average correlation (0.969) than those of AVD (0.847). In 200 total rankings, bAVD misranked 52 and AVD misranked 179 segmentations. Our proposed evaluation measure, bAVD, alleviates AVDs ranking bias making it more suitable for rankings and quality assessment of segmentations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge