Jinjin Cai
Multimodal Remote Sensing Scene Classification Using VLMs and Dual-Cross Attention Networks
Dec 03, 2024
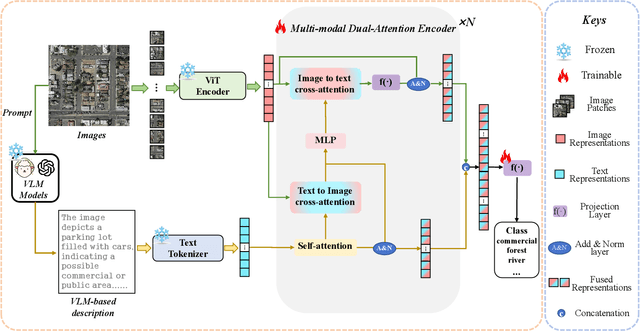
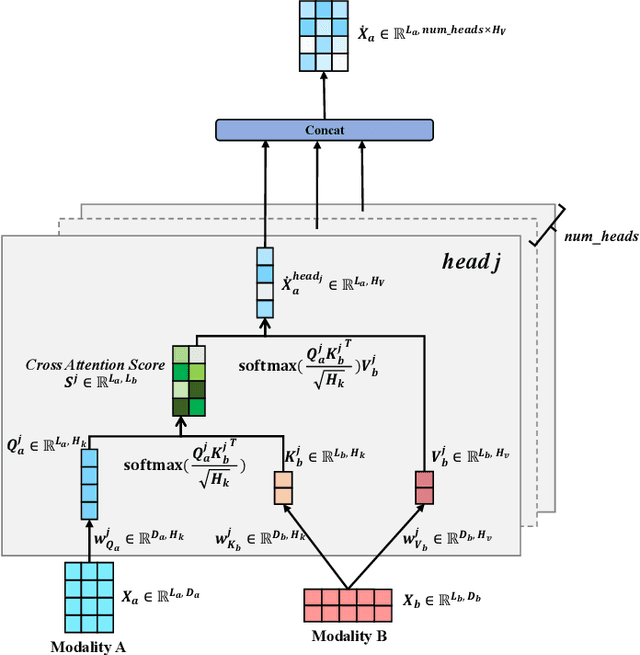
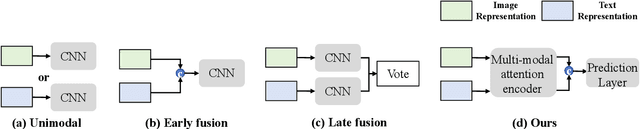
Abstract:Remote sensing scene classification (RSSC) is a critical task with diverse applications in land use and resource management. While unimodal image-based approaches show promise, they often struggle with limitations such as high intra-class variance and inter-class similarity. Incorporating textual information can enhance classification by providing additional context and semantic understanding, but manual text annotation is labor-intensive and costly. In this work, we propose a novel RSSC framework that integrates text descriptions generated by large vision-language models (VLMs) as an auxiliary modality without incurring expensive manual annotation costs. To fully leverage the latent complementarities between visual and textual data, we propose a dual cross-attention-based network to fuse these modalities into a unified representation. Extensive experiments with both quantitative and qualitative evaluation across five RSSC datasets demonstrate that our framework consistently outperforms baseline models. We also verify the effectiveness of VLM-generated text descriptions compared to human-annotated descriptions. Additionally, we design a zero-shot classification scenario to show that the learned multimodal representation can be effectively utilized for unseen class classification. This research opens new opportunities for leveraging textual information in RSSC tasks and provides a promising multimodal fusion structure, offering insights and inspiration for future studies. Code is available at: https://github.com/CJR7/MultiAtt-RSSC
AuD-Former: A Hierarchical Transformer Network for Multimodal Audio-Based Disease Prediction
Oct 11, 2024



Abstract:Audio-based disease prediction is emerging as a promising supplement to traditional medical diagnosis methods, facilitating early, convenient, and non-invasive disease detection and prevention. Multimodal fusion, which integrates features from various domains within or across bio-acoustic modalities, has proven effective in enhancing diagnostic performance. However, most existing methods in the field employ unilateral fusion strategies that focus solely on either intra-modal or inter-modal fusion. This approach limits the full exploitation of the complementary nature of diverse acoustic feature domains and bio-acoustic modalities. Additionally, the inadequate and isolated exploration of latent dependencies within modality-specific and modality-shared spaces curtails their capacity to manage the inherent heterogeneity in multimodal data. To fill these gaps, we propose AuD-Former, a hierarchical transformer network designed for general multimodal audio-based disease prediction. Specifically, we seamlessly integrate intra-modal and inter-modal fusion in a hierarchical manner and proficiently encode the necessary intra-modal and inter-modal complementary correlations, respectively. Comprehensive experiments demonstrate that AuD-Former achieves state-of-the-art performance in predicting three diseases: COVID-19, Parkinson's disease, and pathological dysarthria, showcasing its promising potential in a broad context of audio-based disease prediction tasks. Additionally, extensive ablation studies and qualitative analyses highlight the significant benefits of each main component within our model.
Discovering COVID-19 Coughing and Breathing Patterns from Unlabeled Data Using Contrastive Learning with Varying Pre-Training Domains
Jun 02, 2023


Abstract:Rapid discovery of new diseases, such as COVID-19 can enable a timely epidemic response, preventing the large-scale spread and protecting public health. However, limited research efforts have been taken on this problem. In this paper, we propose a contrastive learning-based modeling approach for COVID-19 coughing and breathing pattern discovery from non-COVID coughs. To validate our models, extensive experiments have been conducted using four large audio datasets and one image dataset. We further explore the effects of different factors, such as domain relevance and augmentation order on the pre-trained models. Our results show that the proposed model can effectively distinguish COVID-19 coughing and breathing from unlabeled data and labeled non-COVID coughs with an accuracy of up to 0.81 and 0.86, respectively. Findings from this work will guide future research to detect an outbreak of a new disease early.
* Accepted by Proceedings of INTERSPEECH 2023
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge