Jiangzhuo Chen
Wisdom of the Ensemble: Improving Consistency of Deep Learning Models
Nov 13, 2020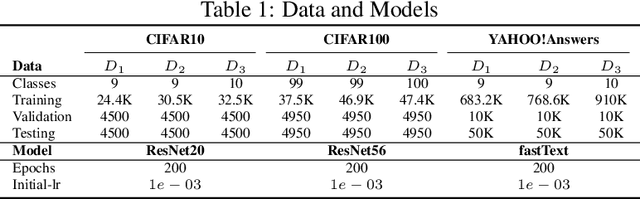
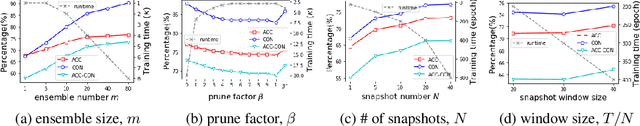
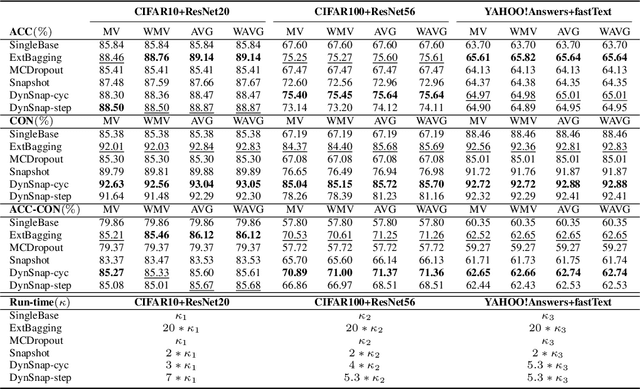
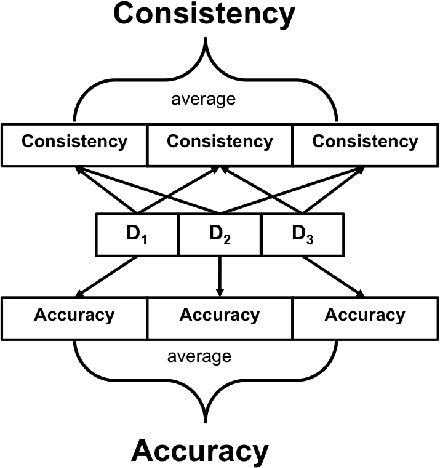
Abstract:Deep learning classifiers are assisting humans in making decisions and hence the user's trust in these models is of paramount importance. Trust is often a function of constant behavior. From an AI model perspective it means given the same input the user would expect the same output, especially for correct outputs, or in other words consistently correct outputs. This paper studies a model behavior in the context of periodic retraining of deployed models where the outputs from successive generations of the models might not agree on the correct labels assigned to the same input. We formally define consistency and correct-consistency of a learning model. We prove that consistency and correct-consistency of an ensemble learner is not less than the average consistency and correct-consistency of individual learners and correct-consistency can be improved with a probability by combining learners with accuracy not less than the average accuracy of ensemble component learners. To validate the theory using three datasets and two state-of-the-art deep learning classifiers we also propose an efficient dynamic snapshot ensemble method and demonstrate its value.
Examining Deep Learning Models with Multiple Data Sources for COVID-19 Forecasting
Oct 27, 2020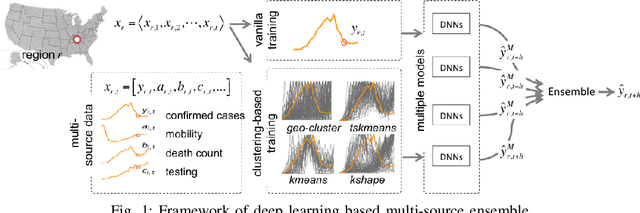
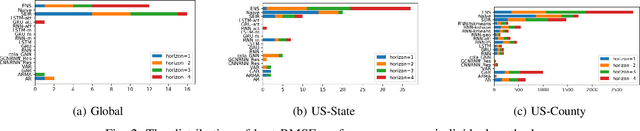
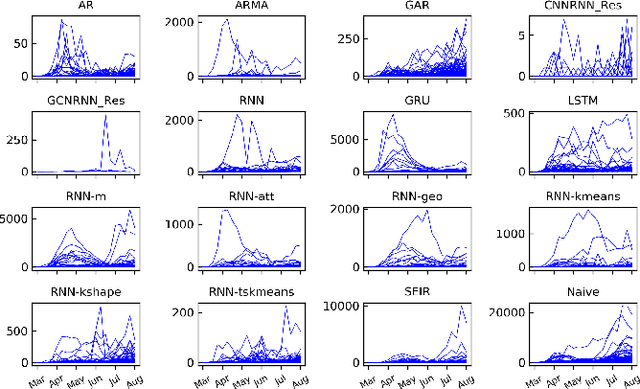
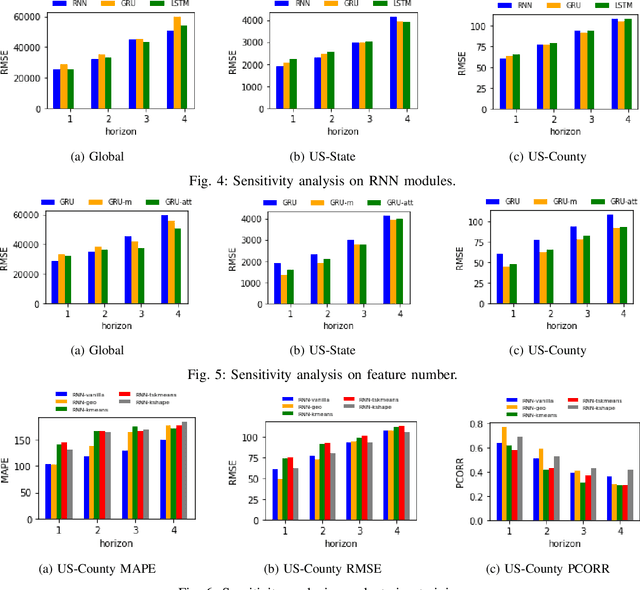
Abstract:The COVID-19 pandemic represents the most significant public health disaster since the 1918 influenza pandemic. During pandemics such as COVID-19, timely and reliable spatio-temporal forecasting of epidemic dynamics is crucial. Deep learning-based time series models for forecasting have recently gained popularity and have been successfully used for epidemic forecasting. Here we focus on the design and analysis of deep learning-based models for COVID-19 forecasting. We implement multiple recurrent neural network-based deep learning models and combine them using the stacking ensemble technique. In order to incorporate the effects of multiple factors in COVID-19 spread, we consider multiple sources such as COVID-19 testing data and human mobility data for better predictions. To overcome the sparsity of training data and to address the dynamic correlation of the disease, we propose clustering-based training for high-resolution forecasting. The methods help us to identify the similar trends of certain groups of regions due to various spatio-temporal effects. We examine the proposed method for forecasting weekly COVID-19 new confirmed cases at county-, state-, and country-level. A comprehensive comparison between different time series models in COVID-19 context is conducted and analyzed. The results show that simple deep learning models can achieve comparable or better performance when compared with more complicated models. We are currently integrating our methods as a part of our weekly forecasts that we provide state and federal authorities.
TDEFSI: Theory Guided Deep Learning Based Epidemic Forecasting with Synthetic Information
Jan 28, 2020
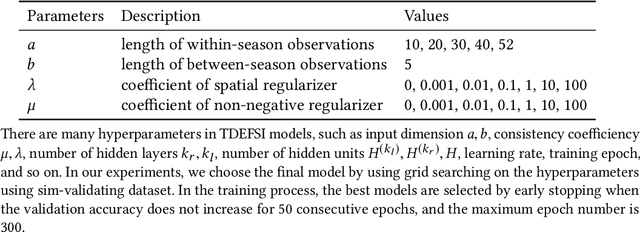
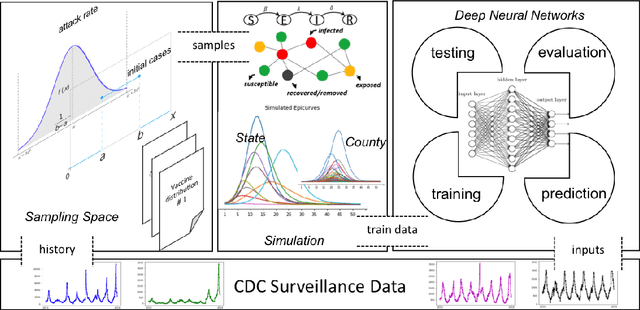
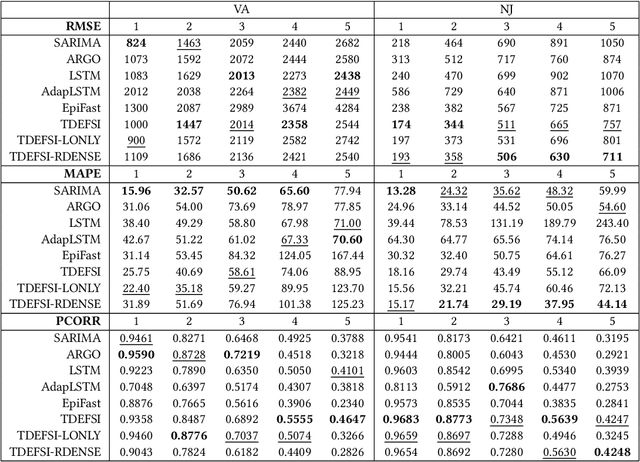
Abstract:Influenza-like illness (ILI) places a heavy social and economic burden on our society. Traditionally, ILI surveillance data is updated weekly and provided at a spatially coarse resolution. Producing timely and reliable high-resolution spatiotemporal forecasts for ILI is crucial for local preparedness and optimal interventions. We present TDEFSI (Theory Guided Deep Learning Based Epidemic Forecasting with Synthetic Information), an epidemic forecasting framework that integrates the strengths of deep neural networks and high-resolution simulations of epidemic processes over networks. TDEFSI yields accurate high-resolution spatiotemporal forecasts using low-resolution time series data. During the training phase, TDEFSI uses high-resolution simulations of epidemics that explicitly model spatial and social heterogeneity inherent in urban regions as one component of training data. We train a two-branch recurrent neural network model to take both within-season and between-season low-resolution observations as features, and output high-resolution detailed forecasts. The resulting forecasts are not just driven by observed data but also capture the intricate social, demographic and geographic attributes of specific urban regions and mathematical theories of disease propagation over networks. We focus on forecasting the incidence of ILI and evaluate TDEFSI's performance using synthetic and real-world testing datasets at the state and county levels in the USA. The results show that, at the state level, our method achieves comparable/better performance than several state-of-the-art methods. At the county level, TDEFSI outperforms the other methods. The proposed method can be applied to other infectious diseases as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge