TDEFSI: Theory Guided Deep Learning Based Epidemic Forecasting with Synthetic Information
Paper and Code
Jan 28, 2020
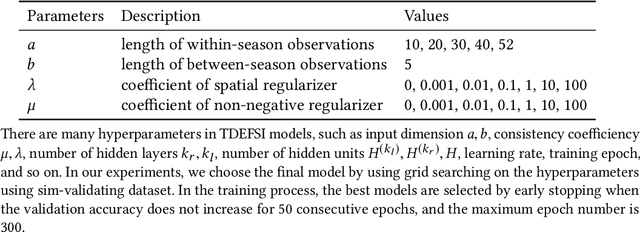
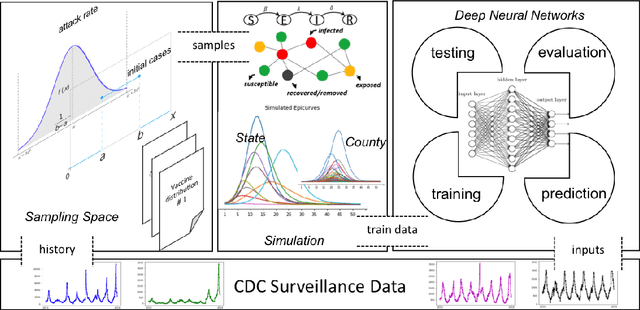
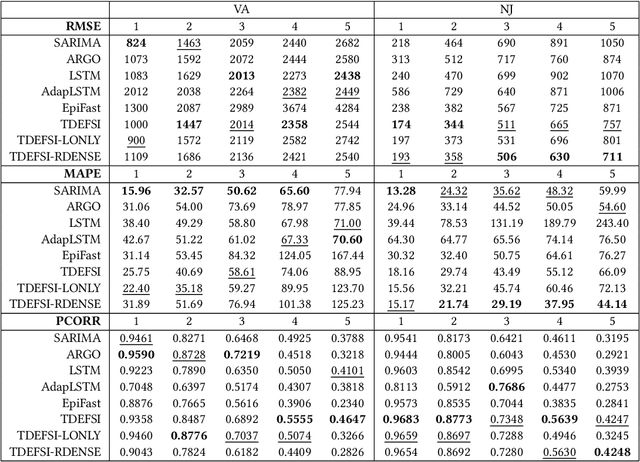
Influenza-like illness (ILI) places a heavy social and economic burden on our society. Traditionally, ILI surveillance data is updated weekly and provided at a spatially coarse resolution. Producing timely and reliable high-resolution spatiotemporal forecasts for ILI is crucial for local preparedness and optimal interventions. We present TDEFSI (Theory Guided Deep Learning Based Epidemic Forecasting with Synthetic Information), an epidemic forecasting framework that integrates the strengths of deep neural networks and high-resolution simulations of epidemic processes over networks. TDEFSI yields accurate high-resolution spatiotemporal forecasts using low-resolution time series data. During the training phase, TDEFSI uses high-resolution simulations of epidemics that explicitly model spatial and social heterogeneity inherent in urban regions as one component of training data. We train a two-branch recurrent neural network model to take both within-season and between-season low-resolution observations as features, and output high-resolution detailed forecasts. The resulting forecasts are not just driven by observed data but also capture the intricate social, demographic and geographic attributes of specific urban regions and mathematical theories of disease propagation over networks. We focus on forecasting the incidence of ILI and evaluate TDEFSI's performance using synthetic and real-world testing datasets at the state and county levels in the USA. The results show that, at the state level, our method achieves comparable/better performance than several state-of-the-art methods. At the county level, TDEFSI outperforms the other methods. The proposed method can be applied to other infectious diseases as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge