Jiabei Zhu
Local Conditional Neural Fields for Versatile and Generalizable Large-Scale Reconstructions in Computational Imaging
Jul 22, 2023



Abstract:Deep learning has transformed computational imaging, but traditional pixel-based representations limit their ability to capture continuous, multiscale details of objects. Here we introduce a novel Local Conditional Neural Fields (LCNF) framework, leveraging a continuous implicit neural representation to address this limitation. LCNF enables flexible object representation and facilitates the reconstruction of multiscale information. We demonstrate the capabilities of LCNF in solving the highly ill-posed inverse problem in Fourier ptychographic microscopy (FPM) with multiplexed measurements, achieving robust, scalable, and generalizable large-scale phase retrieval. Unlike traditional neural fields frameworks, LCNF incorporates a local conditional representation that promotes model generalization, learning multiscale information, and efficient processing of large-scale imaging data. By combining an encoder and a decoder conditioned on a learned latent vector, LCNF achieves versatile continuous-domain super-resolution image reconstruction. We demonstrate accurate reconstruction of wide field-of-view, high-resolution phase images using only a few multiplexed measurements. LCNF robustly captures the continuous object priors and eliminates various phase artifacts, even when it is trained on imperfect datasets. The framework exhibits strong generalization, reconstructing diverse objects even with limited training data. Furthermore, LCNF can be trained on a physics simulator using natural images and successfully applied to experimental measurements on biological samples. Our results highlight the potential of LCNF for solving large-scale inverse problems in computational imaging, with broad applicability in various deep-learning-based techniques.
High-fidelity intensity diffraction tomography with a non-paraxial multiple-scattering model
Jul 13, 2022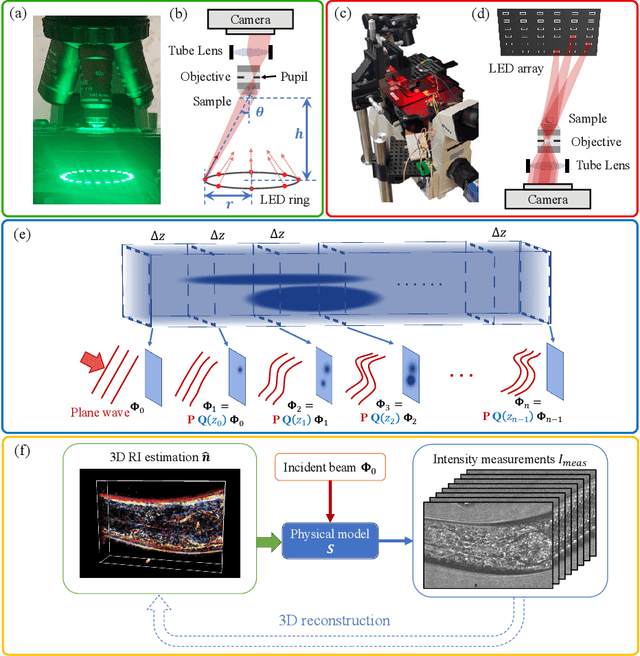
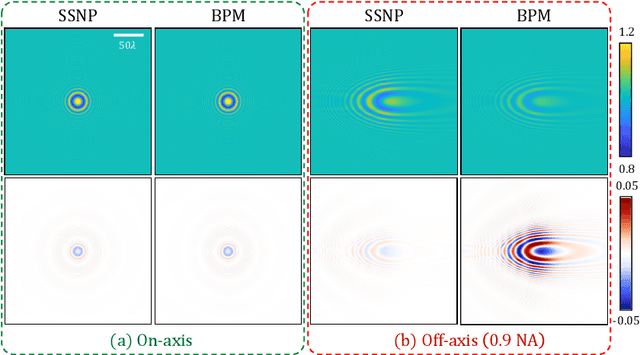
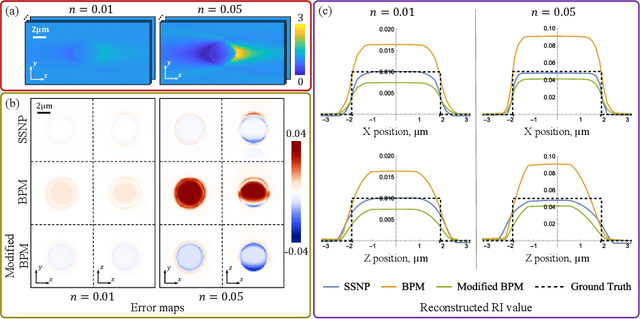
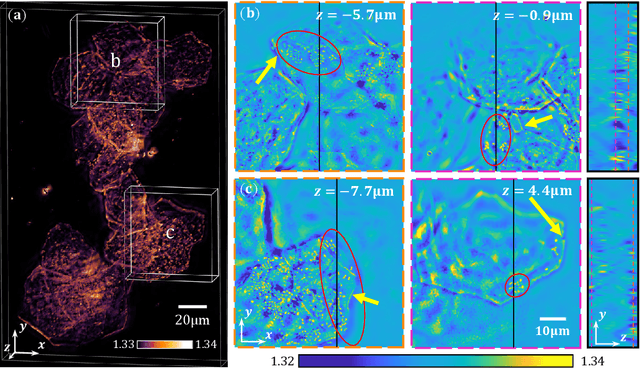
Abstract:We propose a novel intensity diffraction tomography (IDT) reconstruction algorithm based on the split-step non-paraxial (SSNP) model for recovering the 3D refractive index (RI) distribution of multiple-scattering biological samples. High-quality IDT reconstruction requires high-angle illumination to encode both low- and high- spatial frequency information of the 3D biological sample. We show that our SSNP model can more accurately compute multiple scattering from high-angle illumination compared to paraxial approximation-based multiple-scattering models. We apply this SSNP model to both sequential and multiplexed IDT techniques. We develop a unified reconstruction algorithm for both IDT modalities that is highly computationally efficient and is implemented by a modular automatic differentiation framework. We demonstrate the capability of our reconstruction algorithm on both weakly scattering buccal epithelial cells and strongly scattering live $\textit{C. elegans}$ worms and live $\textit{C. elegans}$ embryos.
Zero-Shot Learning of Continuous 3D Refractive Index Maps from Discrete Intensity-Only Measurements
Nov 27, 2021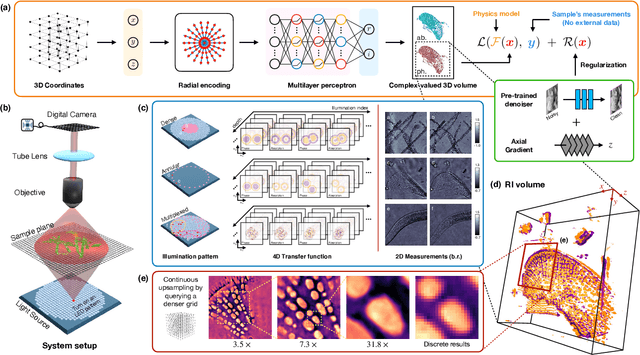

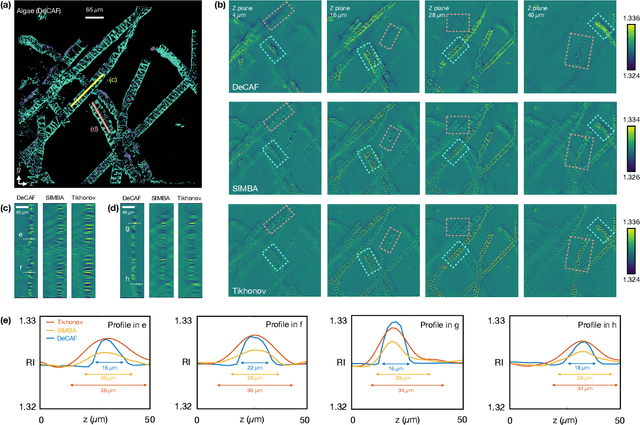
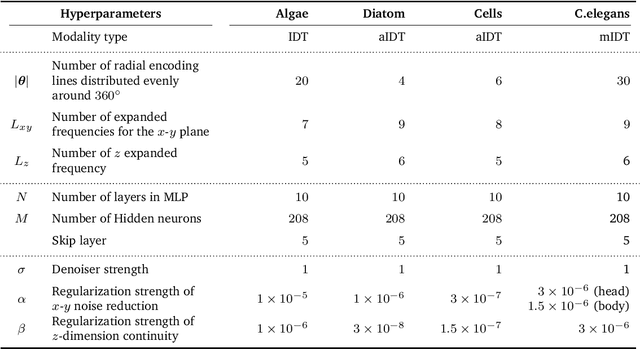
Abstract:Intensity diffraction tomography (IDT) refers to a class of optical microscopy techniques for imaging the 3D refractive index (RI) distribution of a sample from a set of 2D intensity-only measurements. The reconstruction of artifact-free RI maps is a fundamental challenge in IDT due to the loss of phase information and the missing cone problem. Neural fields (NF) has recently emerged as a new deep learning (DL) paradigm for learning continuous representations of complex 3D scenes without external training datasets. We present DeCAF as the first NF-based IDT method that can learn a high-quality continuous representation of a RI volume directly from its intensity-only and limited-angle measurements. We show on three different IDT modalities and multiple biological samples that DeCAF can generate high-contrast and artifact-free RI maps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge