Jessica Knight
FlexICL: A Flexible Visual In-context Learning Framework for Elbow and Wrist Ultrasound Segmentation
Oct 30, 2025Abstract:Elbow and wrist fractures are the most common fractures in pediatric populations. Automatic segmentation of musculoskeletal structures in ultrasound (US) can improve diagnostic accuracy and treatment planning. Fractures appear as cortical defects but require expert interpretation. Deep learning (DL) can provide real-time feedback and highlight key structures, helping lightly trained users perform exams more confidently. However, pixel-wise expert annotations for training remain time-consuming and costly. To address this challenge, we propose FlexICL, a novel and flexible in-context learning (ICL) framework for segmenting bony regions in US images. We apply it to an intra-video segmentation setting, where experts annotate only a small subset of frames, and the model segments unseen frames. We systematically investigate various image concatenation techniques and training strategies for visual ICL and introduce novel concatenation methods that significantly enhance model performance with limited labeled data. By integrating multiple augmentation strategies, FlexICL achieves robust segmentation performance across four wrist and elbow US datasets while requiring only 5% of the training images. It outperforms state-of-the-art visual ICL models like Painter, MAE-VQGAN, and conventional segmentation models like U-Net and TransUNet by 1-27% Dice coefficient on 1,252 US sweeps. These initial results highlight the potential of FlexICL as an efficient and scalable solution for US image segmentation well suited for medical imaging use cases where labeled data is scarce.
A Simple Framework Uniting Visual In-context Learning with Masked Image Modeling to Improve Ultrasound Segmentation
Mar 08, 2024



Abstract:Conventional deep learning models deal with images one-by-one, requiring costly and time-consuming expert labeling in the field of medical imaging, and domain-specific restriction limits model generalizability. Visual in-context learning (ICL) is a new and exciting area of research in computer vision. Unlike conventional deep learning, ICL emphasizes the model's ability to adapt to new tasks based on given examples quickly. Inspired by MAE-VQGAN, we proposed a new simple visual ICL method called SimICL, combining visual ICL pairing images with masked image modeling (MIM) designed for self-supervised learning. We validated our method on bony structures segmentation in a wrist ultrasound (US) dataset with limited annotations, where the clinical objective was to segment bony structures to help with further fracture detection. We used a test set containing 3822 images from 18 patients for bony region segmentation. SimICL achieved an remarkably high Dice coeffient (DC) of 0.96 and Jaccard Index (IoU) of 0.92, surpassing state-of-the-art segmentation and visual ICL models (a maximum DC 0.86 and IoU 0.76), with SimICL DC and IoU increasing up to 0.10 and 0.16. This remarkably high agreement with limited manual annotations indicates SimICL could be used for training AI models even on small US datasets. This could dramatically decrease the human expert time required for image labeling compared to conventional approaches, and enhance the real-world use of AI assistance in US image analysis.
Self-supervised TransUNet for Ultrasound regional segmentation of the distal radius in children
Sep 18, 2023



Abstract:Supervised deep learning offers great promise to automate analysis of medical images from segmentation to diagnosis. However, their performance highly relies on the quality and quantity of the data annotation. Meanwhile, curating large annotated datasets for medical images requires a high level of expertise, which is time-consuming and expensive. Recently, to quench the thirst for large data sets with high-quality annotation, self-supervised learning (SSL) methods using unlabeled domain-specific data, have attracted attention. Therefore, designing an SSL method that relies on minimal quantities of labeled data has far-reaching significance in medical images. This paper investigates the feasibility of deploying the Masked Autoencoder for SSL (SSL-MAE) of TransUNet, for segmenting bony regions from children's wrist ultrasound scans. We found that changing the embedding and loss function in SSL-MAE can produce better downstream results compared to the original SSL-MAE. In addition, we determined that only pretraining TransUNet embedding and encoder with SSL-MAE does not work as well as TransUNet without SSL-MAE pretraining on downstream segmentation tasks.
Physics Driven Domain Specific Transporter Framework with Attention Mechanism for Ultrasound Imaging
Sep 13, 2021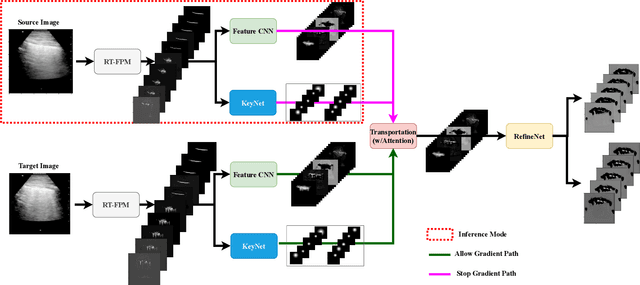
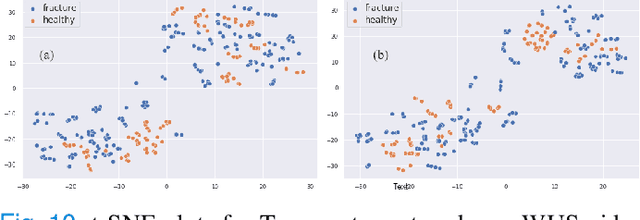
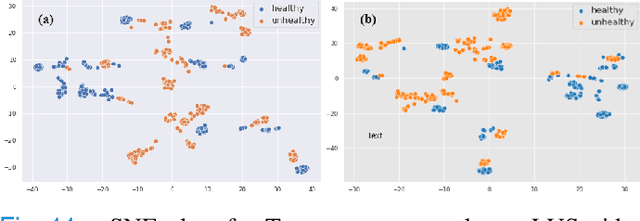
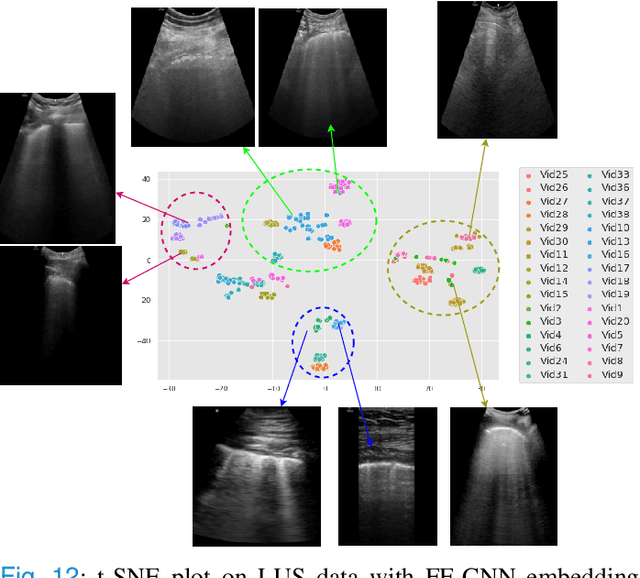
Abstract:Most applications of deep learning techniques in medical imaging are supervised and require a large number of labeled data which is expensive and requires many hours of careful annotation by experts. In this paper, we propose an unsupervised, physics driven domain specific transporter framework with an attention mechanism to identify relevant key points with applications in ultrasound imaging. The proposed framework identifies key points that provide a concise geometric representation highlighting regions with high structural variation in ultrasound videos. We incorporate physics driven domain specific information as a feature probability map and use the radon transform to highlight features in specific orientations. The proposed framework has been trained on130 Lung ultrasound (LUS) videos and 113 Wrist ultrasound (WUS) videos and validated on 100 Lung ultrasound (LUS) videos and 58 Wrist ultrasound (WUS) videos acquired from multiple centers across the globe. Images from both datasets were independently assessed by experts to identify clinically relevant features such as A-lines, B-lines and pleura from LUS and radial metaphysis, radial epiphysis and carpal bones from WUS videos. The key points detected from both datasets showed high sensitivity (LUS = 99\% , WUS = 74\%) in detecting the image landmarks identified by experts. Also, on employing for classification of the given lung image into normal and abnormal classes, the proposed approach, even with no prior training, achieved an average accuracy of 97\% and an average F1-score of 95\% respectively on the task of co-classification with 3 fold cross-validation. With the purely unsupervised nature of the proposed approach, we expect the key point detection approach to increase the applicability of ultrasound in various examination performed in emergency and point of care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge