Jay D. Humphrey
Importance of localized dilatation and distensibility in identifying determinants of thoracic aortic aneurysm with neural operators
Sep 30, 2025Abstract:Thoracic aortic aneurysms (TAAs) arise from diverse mechanical and mechanobiological disruptions to the aortic wall that increase the risk of dissection or rupture. Evidence links TAA development to dysfunctions in the aortic mechanotransduction axis, including loss of elastic fiber integrity and cell-matrix connections. Because distinct insults create different mechanical vulnerabilities, there is a critical need to identify interacting factors that drive progression. Here, we use a finite element framework to generate synthetic TAAs from hundreds of heterogeneous insults spanning varying degrees of elastic fiber damage and impaired mechanosensing. From these simulations, we construct spatial maps of localized dilatation and distensibility to train neural networks that predict the initiating combined insult. We compare several architectures (Deep Operator Networks, UNets, and Laplace Neural Operators) and multiple input data formats to define a standard for future subject-specific modeling. We also quantify predictive performance when networks are trained using only geometric data (dilatation) versus both geometric and mechanical data (dilatation plus distensibility). Across all networks, prediction errors are significantly higher when trained on dilatation alone, underscoring the added value of distensibility information. Among the tested models, UNet consistently provides the highest accuracy across all data formats. These findings highlight the importance of acquiring full-field measurements of both dilatation and distensibility in TAA assessment to reveal the mechanobiological drivers of disease and support the development of personalized treatment strategies.
A Generative Modeling Framework for Inferring Families of Biomechanical Constitutive Laws in Data-Sparse Regimes
May 04, 2023Abstract:Quantifying biomechanical properties of the human vasculature could deepen our understanding of cardiovascular diseases. Standard nonlinear regression in constitutive modeling requires considerable high-quality data and an explicit form of the constitutive model as prior knowledge. By contrast, we propose a novel approach that combines generative deep learning with Bayesian inference to efficiently infer families of constitutive relationships in data-sparse regimes. Inspired by the concept of functional priors, we develop a generative adversarial network (GAN) that incorporates a neural operator as the generator and a fully-connected neural network as the discriminator. The generator takes a vector of noise conditioned on measurement data as input and yields the predicted constitutive relationship, which is scrutinized by the discriminator in the following step. We demonstrate that this framework can accurately estimate means and standard deviations of the constitutive relationships of the murine aorta using data collected either from model-generated synthetic data or ex vivo experiments for mice with genetic deficiencies. In addition, the framework learns priors of constitutive models without explicitly knowing their functional form, providing a new model-agnostic approach to learning hidden constitutive behaviors from data.
G2Φnet: Relating Genotype and Biomechanical Phenotype of Tissues with Deep Learning
Aug 21, 2022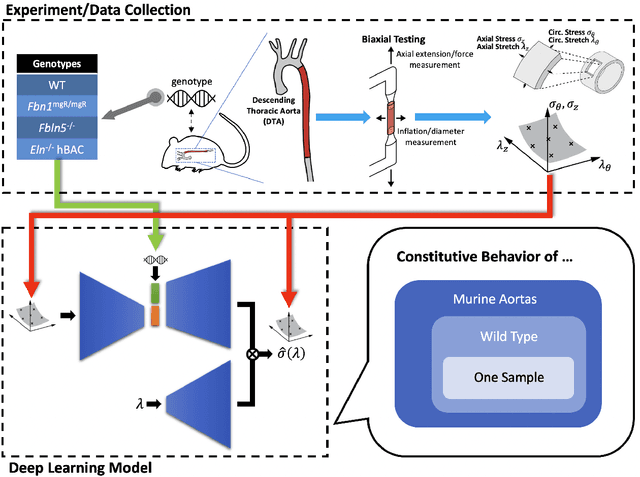
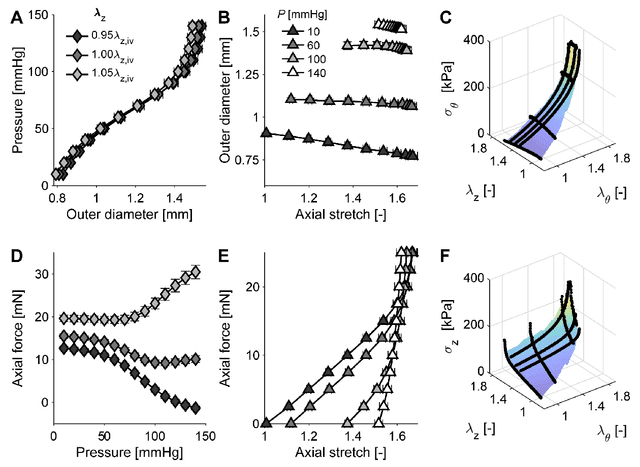
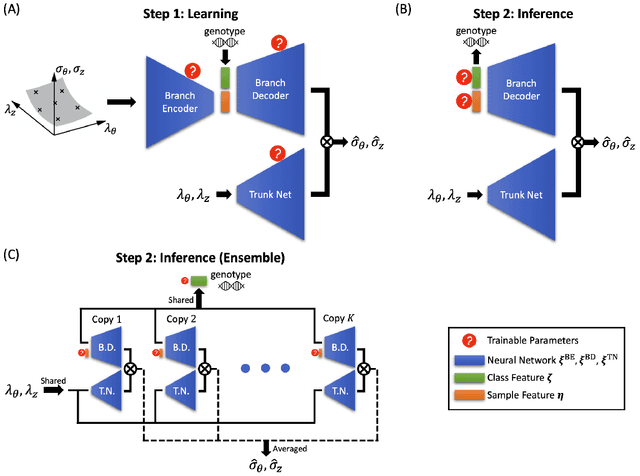
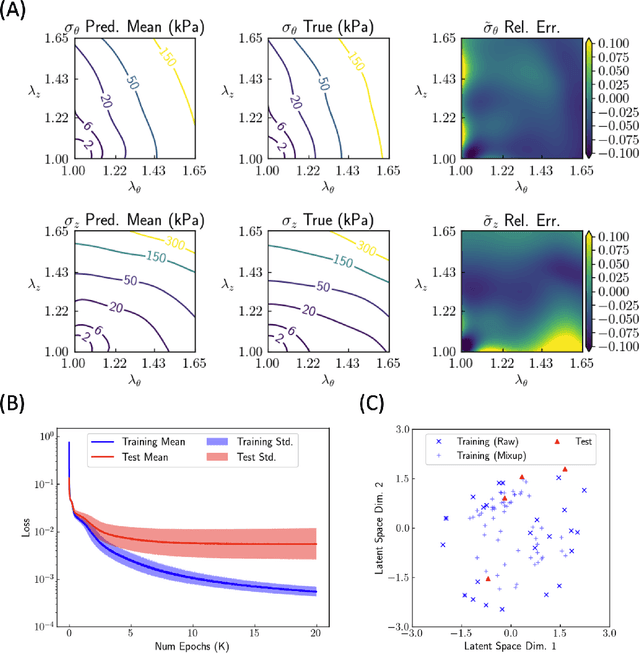
Abstract:Many genetic mutations adversely affect the structure and function of load-bearing soft tissues, with clinical sequelae often responsible for disability or death. Parallel advances in genetics and histomechanical characterization provide significant insight into these conditions, but there remains a pressing need to integrate such information. We present a novel genotype-to-biomechanical-phenotype neural network (G2{\Phi}net) for characterizing and classifying biomechanical properties of soft tissues, which serve as important functional readouts of tissue health or disease. We illustrate the utility of our approach by inferring the nonlinear, genotype-dependent constitutive behavior of the aorta for four mouse models involving defects or deficiencies in extracellular constituents. We show that G2{\Phi}net can infer the biomechanical response while simultaneously ascribing the associated genotype correctly by utilizing limited, noisy, and unstructured experimental data. More broadly, G2{\Phi}net provides a powerful method and a paradigm shift for correlating genotype and biomechanical phenotype quantitatively, promising a better understanding of their interplay in biological tissues.
Neural operator learning of heterogeneous mechanobiological insults contributing to aortic aneurysms
May 08, 2022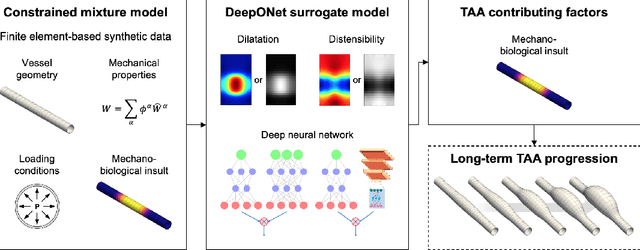
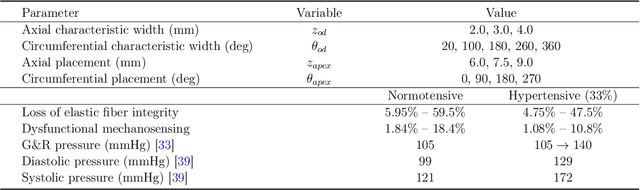
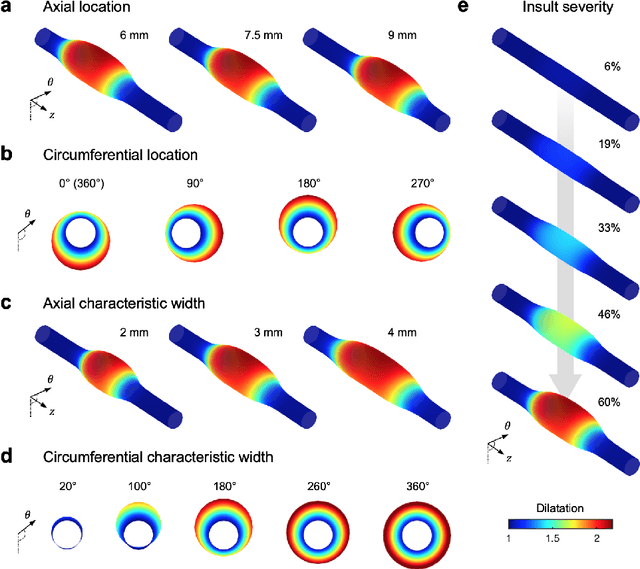

Abstract:Thoracic aortic aneurysm (TAA) is a localized dilatation of the aorta resulting from compromised wall composition, structure, and function, which can lead to life-threatening dissection or rupture. Several genetic mutations and predisposing factors that contribute to TAA have been studied in mouse models to characterize specific changes in aortic microstructure and material properties that result from a wide range of mechanobiological insults. Assessments of TAA progression in vivo is largely limited to measurements of aneurysm size and growth rate. It has been shown that aortic geometry alone is not sufficient to predict the patient-specific progression of TAA but computational modeling of the evolving biomechanics of the aorta could predict future geometry and properties from initiating insults. In this work, we present an integrated framework to train a deep operator network (DeepONet)-based surrogate model to identify contributing factors for TAA by using FE-based datasets of aortic growth and remodeling resulting from prescribed insults. For training data, we investigate multiple types of TAA risk factors and spatial distributions within a constrained mixture model to generate axial--azimuthal maps of aortic dilatation and distensibility. The trained network is then capable of predicting the initial distribution and extent of the insult from a given set of dilatation and distensibility information. Two DeepONet frameworks are proposed, one trained on sparse information and one on full-field grayscale images, to gain insight into a preferred neural operator-based approach. Performance of the surrogate models is evaluated through multiple simulations carried out on insult distributions varying from fusiform to complex. We show that the proposed approach can predict patient-specific mechanobiological insult profile with a high accuracy, particularly when based on full-field images.
Simulating progressive intramural damage leading to aortic dissection using an operator-regression neural network
Aug 25, 2021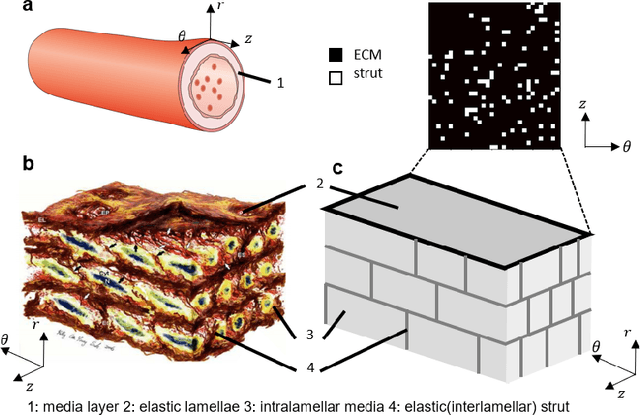
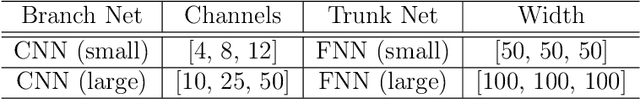


Abstract:Aortic dissection progresses via delamination of the medial layer of the wall. Notwithstanding the complexity of this process, insight has been gleaned by studying in vitro and in silico the progression of dissection driven by quasi-static pressurization of the intramural space by fluid injection, which demonstrates that the differential propensity of dissection can be affected by spatial distributions of structurally significant interlamellar struts that connect adjacent elastic lamellae. In particular, diverse histological microstructures may lead to differential mechanical behavior during dissection, including the pressure--volume relationship of the injected fluid and the displacement field between adjacent lamellae. In this study, we develop a data-driven surrogate model for the delamination process for differential strut distributions using DeepONet, a new operator--regression neural network. The surrogate model is trained to predict the pressure--volume curve of the injected fluid and the damage progression field of the wall given a spatial distribution of struts, with in silico data generated with a phase-field finite element model. The results show that DeepONet can provide accurate predictions for diverse strut distributions, indicating that this composite branch-trunk neural network can effectively extract the underlying functional relationship between distinctive microstructures and their mechanical properties. More broadly, DeepONet can facilitate surrogate model-based analyses to quantify biological variability, improve inverse design, and predict mechanical properties based on multi-modality experimental data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge