Jae-Hyeok Lee
Cosine Similarity Knowledge Distillation for Individual Class Information Transfer
Nov 24, 2023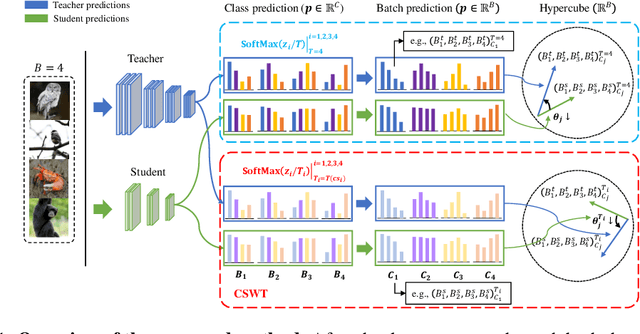

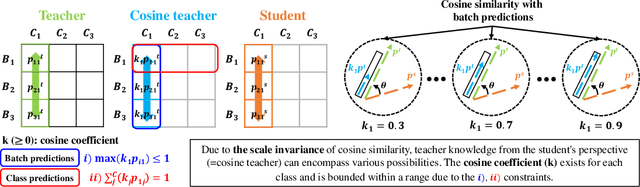

Abstract:Previous logits-based Knowledge Distillation (KD) have utilized predictions about multiple categories within each sample (i.e., class predictions) and have employed Kullback-Leibler (KL) divergence to reduce the discrepancy between the student and teacher predictions. Despite the proliferation of KD techniques, the student model continues to fall short of achieving a similar level as teachers. In response, we introduce a novel and effective KD method capable of achieving results on par with or superior to the teacher models performance. We utilize teacher and student predictions about multiple samples for each category (i.e., batch predictions) and apply cosine similarity, a commonly used technique in Natural Language Processing (NLP) for measuring the resemblance between text embeddings. This metric's inherent scale-invariance property, which relies solely on vector direction and not magnitude, allows the student to dynamically learn from the teacher's knowledge, rather than being bound by a fixed distribution of the teacher's knowledge. Furthermore, we propose a method called cosine similarity weighted temperature (CSWT) to improve the performance. CSWT reduces the temperature scaling in KD when the cosine similarity between the student and teacher models is high, and conversely, it increases the temperature scaling when the cosine similarity is low. This adjustment optimizes the transfer of information from the teacher to the student model. Extensive experimental results show that our proposed method serves as a viable alternative to existing methods. We anticipate that this approach will offer valuable insights for future research on model compression.
Feature2Mass: Visual Feature Processing in Latent Space for Realistic Labeled Mass Generation
Sep 17, 2018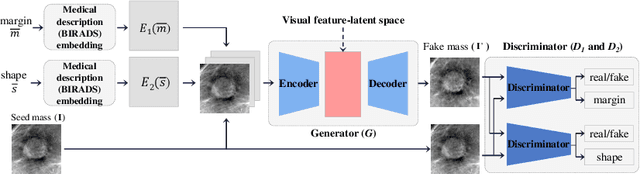
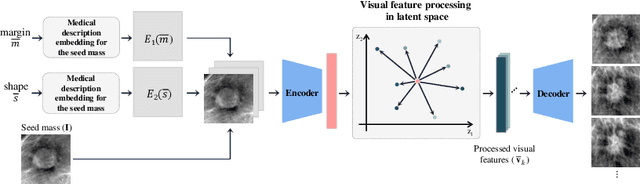
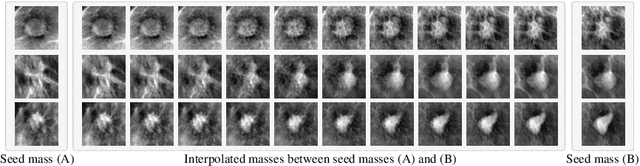
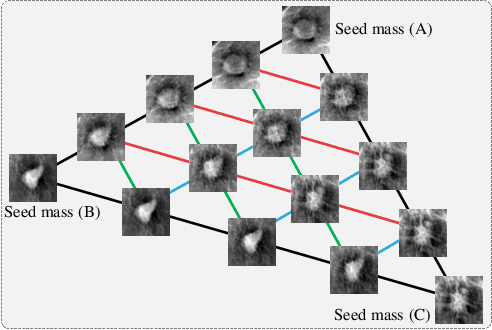
Abstract:This paper deals with a method for generating realistic labeled masses. Recently, there have been many attempts to apply deep learning to various bio-image computing fields including computer-aided detection and diagnosis. In order to learn deep network model to be well-behaved in bio-image computing fields, a lot of labeled data is required. However, in many bioimaging fields, the large-size of labeled dataset is scarcely available. Although a few researches have been dedicated to solving this problem through generative model, there are some problems as follows: 1) The generated bio-image does not seem realistic; 2) the variation of generated bio-image is limited; and 3) additional label annotation task is needed. In this study, we propose a realistic labeled bio-image generation method through visual feature processing in latent space. Experimental results have shown that mass images generated by the proposed method were realistic and had wide expression range of targeted mass characteristics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge