Jacobus W. Portegies
Topological degree as a discrete diagnostic for disentanglement, with applications to the $Δ$VAE
Sep 02, 2024Abstract:We investigate the ability of Diffusion Variational Autoencoder ($\Delta$VAE) with unit sphere $\mathcal{S}^2$ as latent space to capture topological and geometrical structure and disentangle latent factors in datasets. For this, we introduce a new diagnostic of disentanglement: namely the topological degree of the encoder, which is a map from the data manifold to the latent space. By using tools from homology theory, we derive and implement an algorithm that computes this degree. We use the algorithm to compute the degree of the encoder of models that result from the training procedure. Our experimental results show that the $\Delta$VAE achieves relatively small LSBD scores, and that regardless of the degree after initialization, the degree of the encoder after training becomes $-1$ or $+1$, which implies that the resulting encoder is at least homotopic to a homeomorphism.
Neural Langevin Dynamics: towards interpretable Neural Stochastic Differential Equations
Nov 17, 2022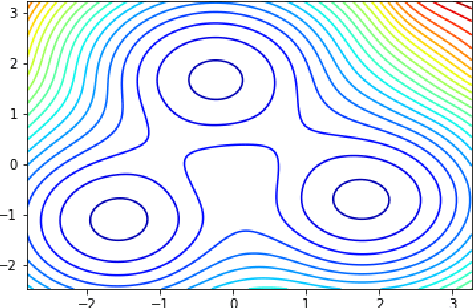
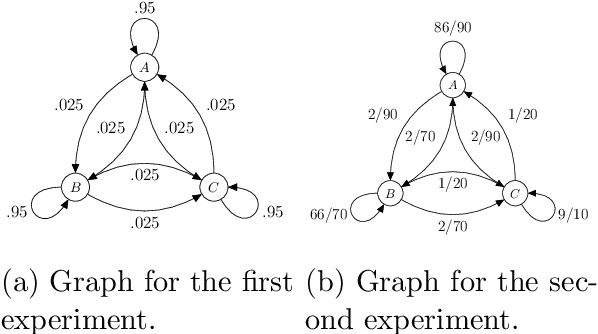
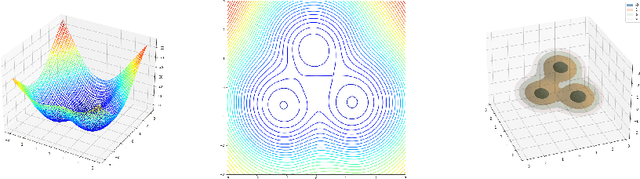
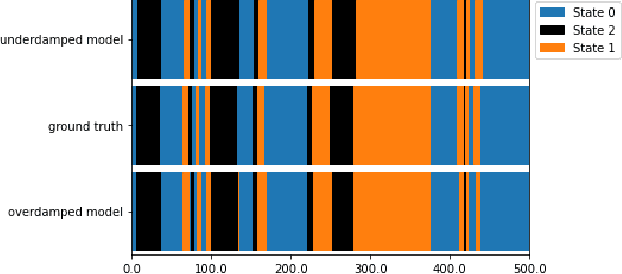
Abstract:Neural Stochastic Differential Equations (NSDE) have been trained as both Variational Autoencoders, and as GANs. However, the resulting Stochastic Differential Equations can be hard to interpret or analyse due to the generic nature of the drift and diffusion fields. By restricting our NSDE to be of the form of Langevin dynamics, and training it as a VAE, we obtain NSDEs that lend themselves to more elaborate analysis and to a wider range of visualisation techniques than a generic NSDE. More specifically, we obtain an energy landscape, the minima of which are in one-to-one correspondence with latent states underlying the used data. This not only allows us to detect states underlying the data dynamics in an unsupervised manner, but also to infer the distribution of time spent in each state according to the learned SDE. More in general, restricting an NSDE to Langevin dynamics enables the use of a large set of tools from computational molecular dynamics for the analysis of the obtained results.
Universal Approximation in Dropout Neural Networks
Dec 18, 2020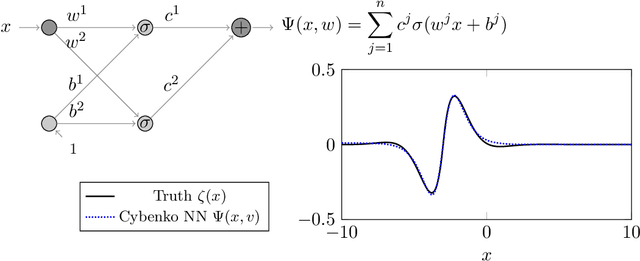
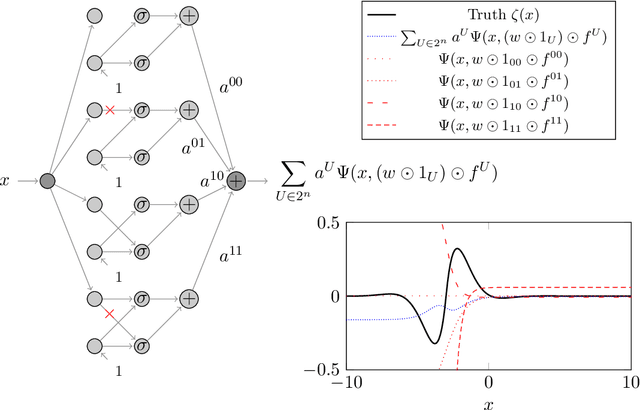
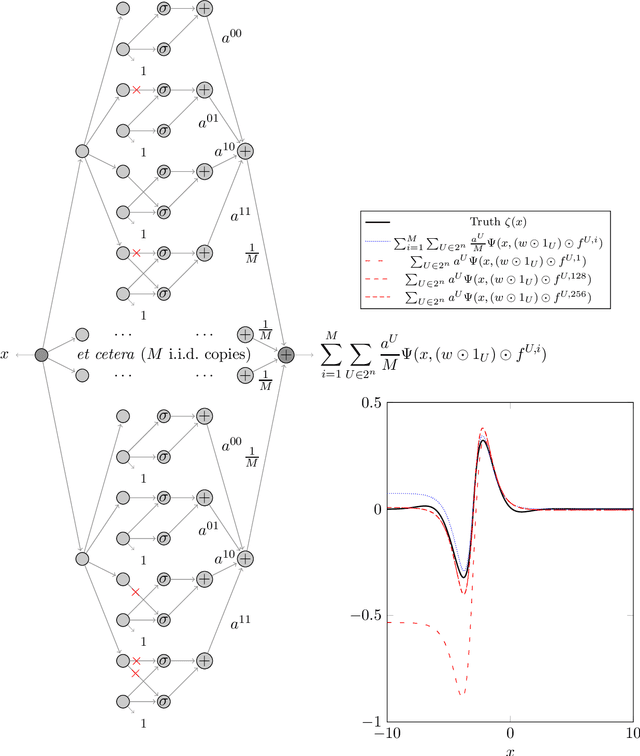
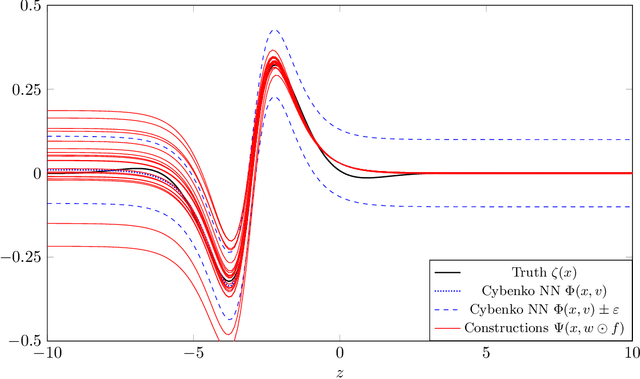
Abstract:We prove two universal approximation theorems for a range of dropout neural networks. These are feed-forward neural networks in which each edge is given a random $\{0,1\}$-valued filter, that have two modes of operation: in the first each edge output is multiplied by its random filter, resulting in a random output, while in the second each edge output is multiplied by the expectation of its filter, leading to a deterministic output. It is common to use the random mode during training and the deterministic mode during testing and prediction. Both theorems are of the following form: Given a function to approximate and a threshold $\varepsilon>0$, there exists a dropout network that is $\varepsilon$-close in probability and in $L^q$. The first theorem applies to dropout networks in the random mode. It assumes little on the activation function, applies to a wide class of networks, and can even be applied to approximation schemes other than neural networks. The core is an algebraic property that shows that deterministic networks can be exactly matched in expectation by random networks. The second theorem makes stronger assumptions and gives a stronger result. Given a function to approximate, it provides existence of a network that approximates in both modes simultaneously. Proof components are a recursive replacement of edges by independent copies, and a special first-layer replacement that couples the resulting larger network to the input. The functions to be approximated are assumed to be elements of general normed spaces, and the approximations are measured in the corresponding norms. The networks are constructed explicitly. Because of the different methods of proof, the two results give independent insight into the approximation properties of random dropout networks. With this, we establish that dropout neural networks broadly satisfy a universal-approximation property.
Quantifying and Learning Disentangled Representations with Limited Supervision
Nov 26, 2020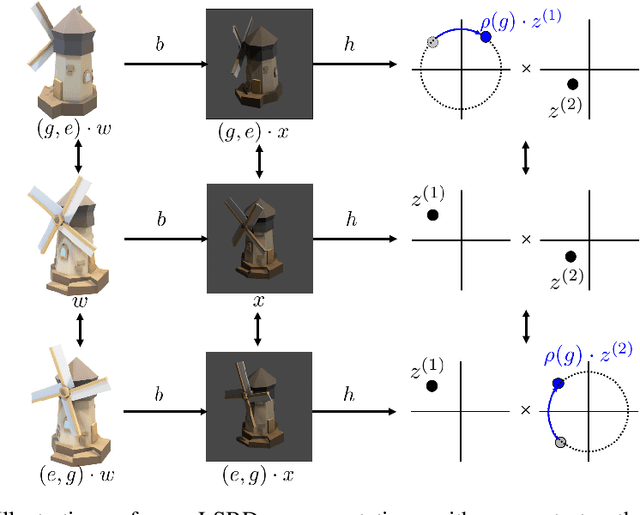
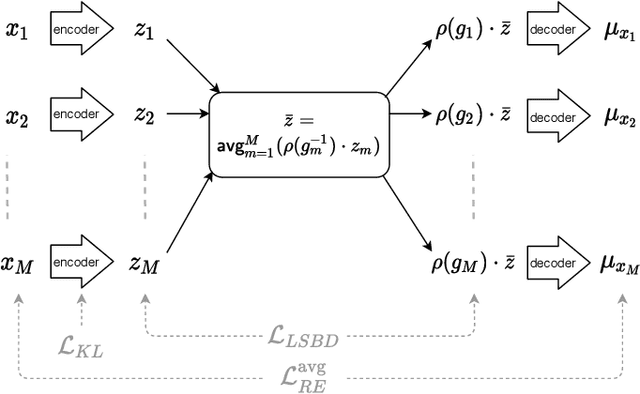

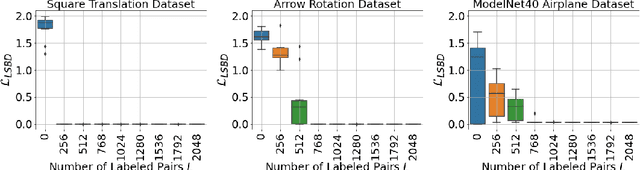
Abstract:Learning low-dimensional representations that disentangle the underlying factors of variation in data has been posited as an important step towards interpretable machine learning with good generalization. To address the fact that there is no consensus on what disentanglement entails, Higgins et al. (2018) propose a formal definition for Linear Symmetry-Based Disentanglement, or LSBD, arguing that underlying real-world transformations give exploitable structure to data. Although several works focus on learning LSBD representations, such methods require supervision on the underlying transformations for the entire dataset, and cannot deal with unlabeled data. Moreover, none of these works provide a metric to quantify LSBD. We propose a metric to quantify LSBD representations that is easy to compute under certain well-defined assumptions. Furthermore, we present a method that can leverage unlabeled data, such that LSBD representations can be learned with limited supervision on transformations. Using our LSBD metric, our results show that limited supervision is indeed sufficient to learn LSBD representations.
A Metric for Linear Symmetry-Based Disentanglement
Nov 26, 2020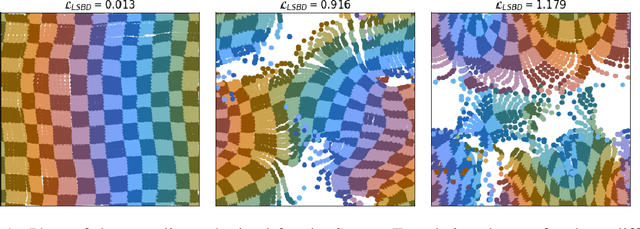

Abstract:The definition of Linear Symmetry-Based Disentanglement (LSBD) proposed by (Higgins et al., 2018) outlines the properties that should characterize a disentangled representation that captures the symmetries of data. However, it is not clear how to measure the degree to which a data representation fulfills these properties. We propose a metric for the evaluation of the level of LSBD that a data representation achieves. We provide a practical method to evaluate this metric and use it to evaluate the disentanglement of the data representations obtained for three datasets with underlying $SO(2)$ symmetries.
Diffusion Variational Autoencoders
Jan 25, 2019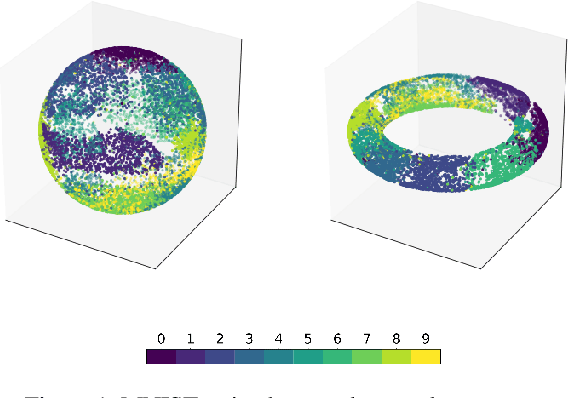
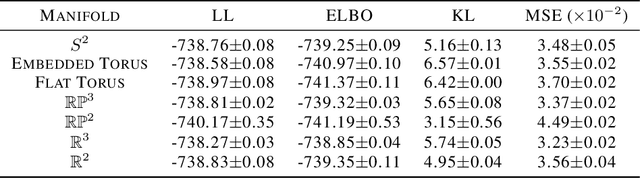
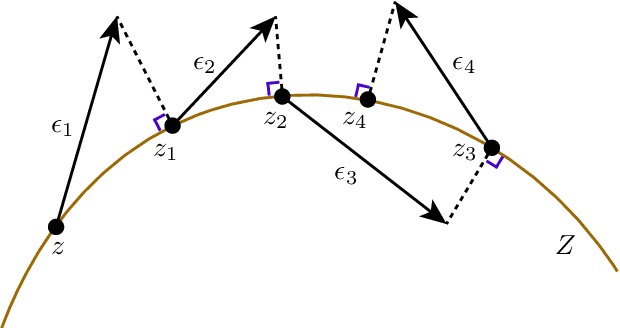
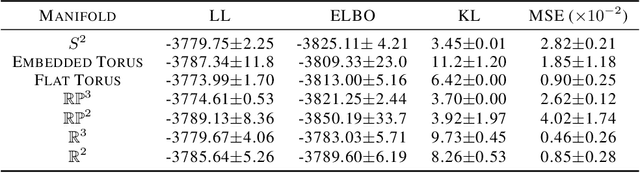
Abstract:A standard Variational Autoencoder, with a Euclidean latent space, is structurally incapable of capturing topological properties of certain datasets. To remove topological obstructions, we introduce Diffusion Variational Autoencoders with arbitrary manifolds as a latent space. A Diffusion Variational Autoencoder uses transition kernels of Brownian motion on the manifold. In particular, it uses properties of the Brownian motion to implement the reparametrization trick and fast approximations to the KL divergence. We show that the Diffusion Variational Autoencoder is capable of capturing topological properties of synthetic datasets. Additionally, we train MNIST on spheres, tori, projective spaces, SO(3), and a torus embedded in R3. Although a natural dataset like MNIST does not have latent variables with a clear-cut topological structure, training it on a manifold can still highlight topological and geometrical properties.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge