Ivana Drobnjak
Towards Multimodal Representation Learning in Paediatric Kidney Disease
Nov 17, 2025


Abstract:Paediatric kidney disease varies widely in its presentation and progression, which calls for continuous monitoring of renal function. Using electronic health records collected between 2019 and 2025 at Great Ormond Street Hospital, a leading UK paediatric hospital, we explored a temporal modelling approach that integrates longitudinal laboratory sequences with demographic information. A recurrent neural model trained on these data was used to predict whether a child would record an abnormal serum creatinine value within the following thirty days. Framed as a pilot study, this work provides an initial demonstration that simple temporal representations can capture useful patterns in routine paediatric data and lays the groundwork for future multimodal extensions using additional clinical signals and more detailed renal outcomes.
Acquisition-invariant brain MRI segmentation with informative uncertainties
Nov 07, 2021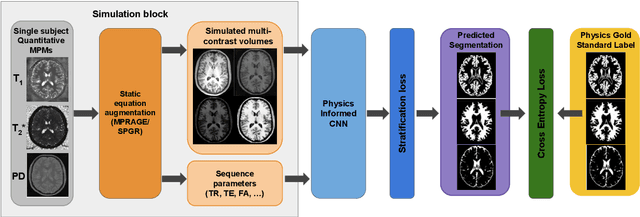
Abstract:Combining multi-site data can strengthen and uncover trends, but is a task that is marred by the influence of site-specific covariates that can bias the data and therefore any downstream analyses. Post-hoc multi-site correction methods exist but have strong assumptions that often do not hold in real-world scenarios. Algorithms should be designed in a way that can account for site-specific effects, such as those that arise from sequence parameter choices, and in instances where generalisation fails, should be able to identify such a failure by means of explicit uncertainty modelling. This body of work showcases such an algorithm, that can become robust to the physics of acquisition in the context of segmentation tasks, while simultaneously modelling uncertainty. We demonstrate that our method not only generalises to complete holdout datasets, preserving segmentation quality, but does so while also accounting for site-specific sequence choices, which also allows it to perform as a harmonisation tool.
The role of MRI physics in brain segmentation CNNs: achieving acquisition invariance and instructive uncertainties
Nov 04, 2021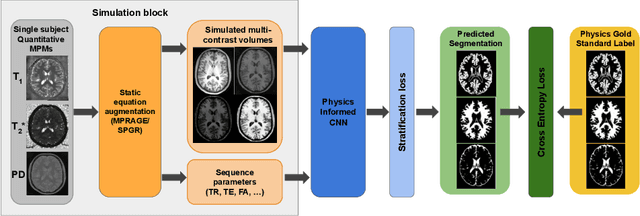
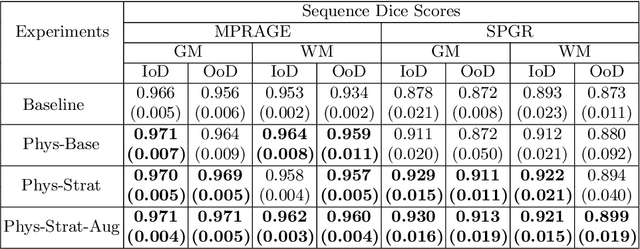
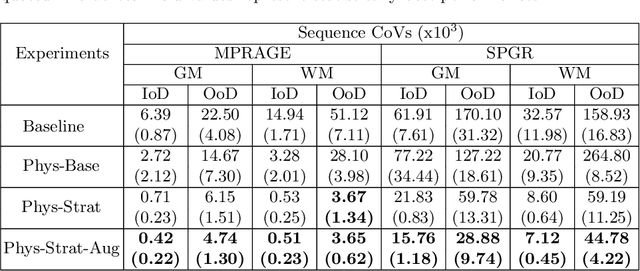
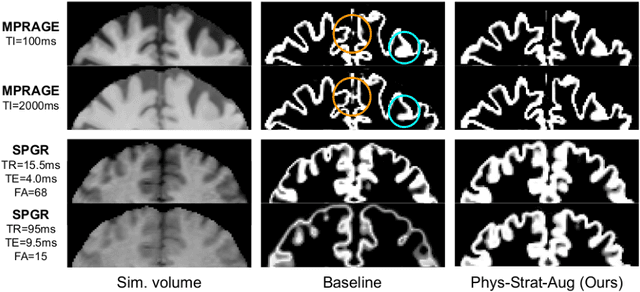
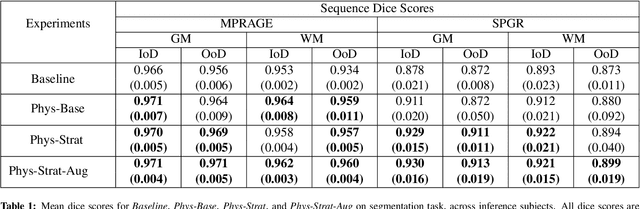
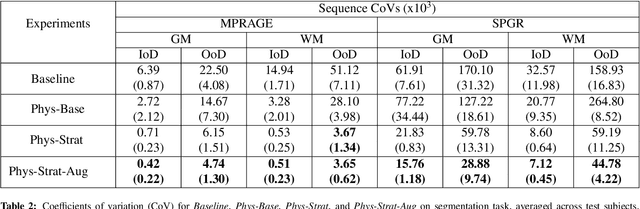
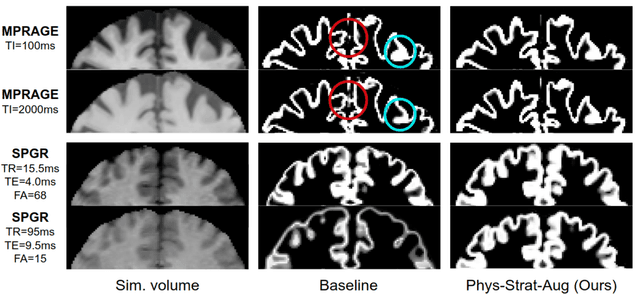
Abstract:Being able to adequately process and combine data arising from different sites is crucial in neuroimaging, but is difficult, owing to site, sequence and acquisition-parameter dependent biases. It is important therefore to design algorithms that are not only robust to images of differing contrasts, but also be able to generalise well to unseen ones, with a quantifiable measure of uncertainty. In this paper we demonstrate the efficacy of a physics-informed, uncertainty-aware, segmentation network that employs augmentation-time MR simulations and homogeneous batch feature stratification to achieve acquisition invariance. We show that the proposed approach also accurately extrapolates to out-of-distribution sequence samples, providing well calibrated volumetric bounds on these. We demonstrate a significant improvement in terms of coefficients of variation, backed by uncertainty based volumetric validation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge