Iain Carmichael
Incorporating intratumoral heterogeneity into weakly-supervised deep learning models via variance pooling
Jun 17, 2022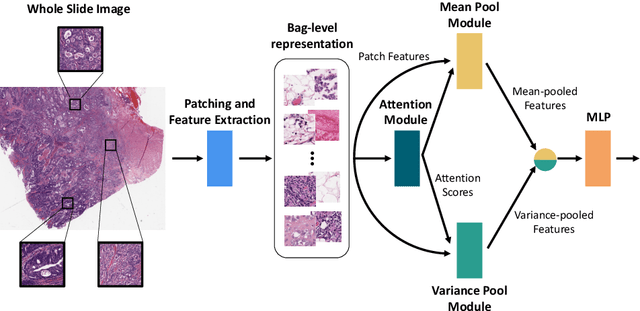
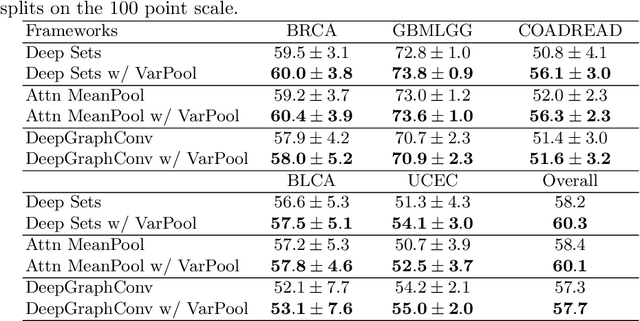
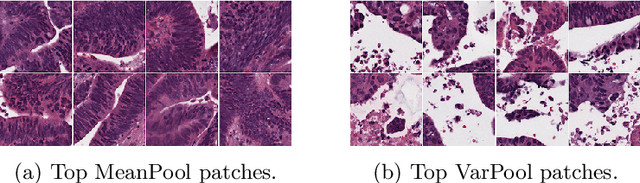
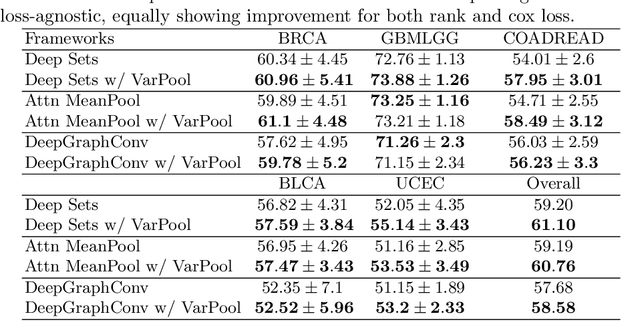
Abstract:Supervised learning tasks such as cancer survival prediction from gigapixel whole slide images (WSIs) are a critical challenge in computational pathology that requires modeling complex features of the tumor microenvironment. These learning tasks are often solved with deep multi-instance learning (MIL) models that do not explicitly capture intratumoral heterogeneity. We develop a novel variance pooling architecture that enables a MIL model to incorporate intratumoral heterogeneity into its predictions. Two interpretability tools based on representative patches are illustrated to probe the biological signals captured by these models. An empirical study with 4,479 gigapixel WSIs from the Cancer Genome Atlas shows that adding variance pooling onto MIL frameworks improves survival prediction performance for five cancer types.
Geometric Insights into Support Vector Machine Behavior using the KKT Conditions
Oct 10, 2018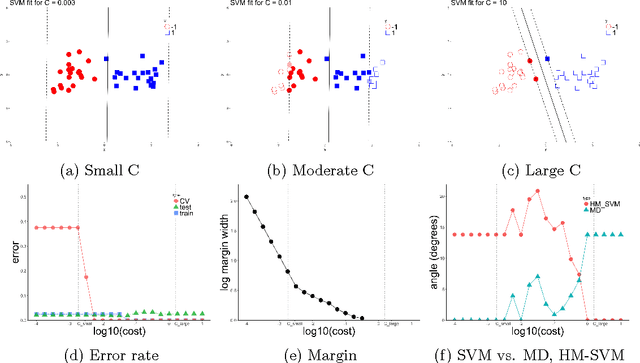
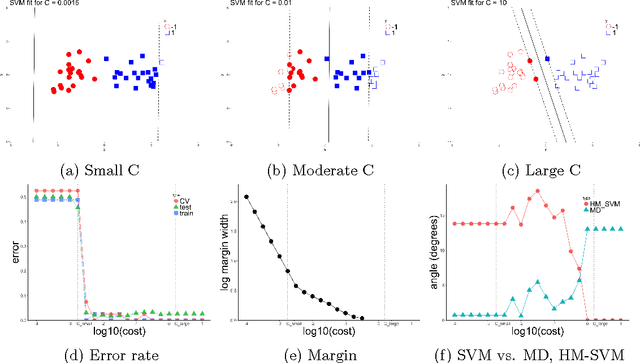
Abstract:The support vector machine (SVM) is a powerful and widely used classification algorithm. This paper uses the Karush-Kuhn-Tucker conditions to provide rigorous mathematical proof for new insights into the behavior of SVM. These insights provide perhaps unexpected relationships between SVM and two other linear classifiers: the mean difference and the maximal data piling direction. For example, we show that in many cases SVM can be viewed as a cropped version of these classifiers. By carefully exploring these connections we show how SVM tuning behavior is affected by characteristics including: balanced vs. unbalanced classes, low vs. high dimension, separable vs. non-separable data. These results provide further insights into tuning SVM via cross-validation by explaining observed pathological behavior and motivating improved cross-validation methodology. Finally, we also provide new results on the geometry of complete data piling directions in high dimensional space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge