Huiye Liu
TransFuse: Fusing Transformers and CNNs for Medical Image Segmentation
Feb 16, 2021



Abstract:U-Net based convolutional neural networks with deep feature representation and skip-connections have significantly boosted the performance of medical image segmentation. In this paper, we study the more challenging problem of improving efficiency in modeling global contexts without losing localization ability for low-level details. TransFuse, a novel two-branch architecture is proposed, which combines Transformers and CNNs in a parallel style. With TransFuse, both global dependency and low-level spatial details can be efficiently captured in a much shallower manner. Besides, a novel fusion technique - BiFusion module is proposed to fuse the multi-level features from each branch. TransFuse achieves the newest state-of-the-arts on polyp segmentation task, with 20\% fewer parameters and the fastest inference speed at about 98.7 FPS.
Improve Model Generalization and Robustness to Dataset Bias with Bias-regularized Learning and Domain-guided Augmentation
Nov 13, 2019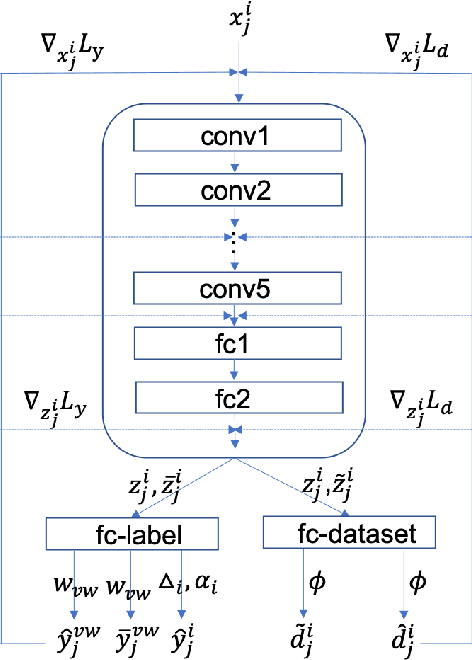
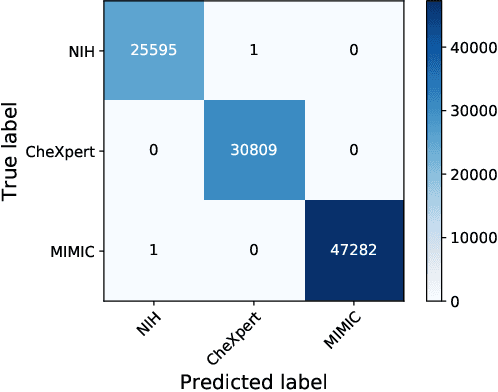
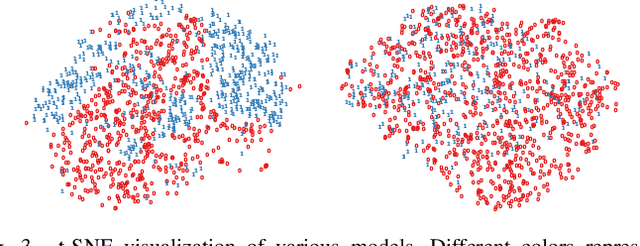
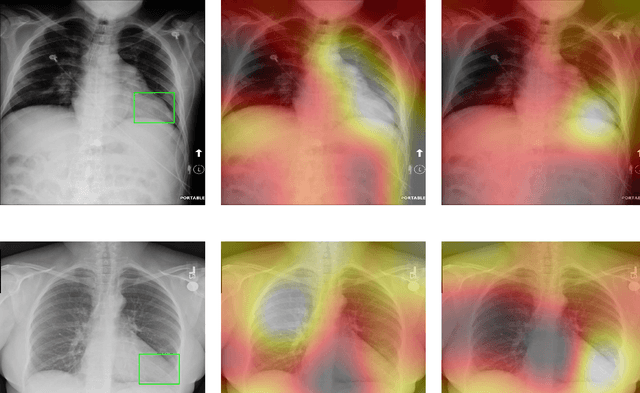
Abstract:Deep Learning has thrived on the emergence of biomedical big data. However, medical datasets acquired at different institutions have inherent bias caused by various confounding factors such as operation policies, machine protocols, treatment preference and etc. As the result, models trained on one dataset, regardless of volume, cannot be confidently utilized for the others. In this study, we investigated model robustness to dataset bias using three large-scale Chest X-ray datasets: first, we assessed the dataset bias using vanilla training baseline; second, we proposed a novel multi-source domain generalization model by (a) designing a new bias-regularized loss function; and (b) synthesizing new data for domain augmentation. We showed that our model significantly outperformed the baseline and other approaches on data from unseen domain in terms of accuracy and various bias measures, without retraining or finetuning. Our method is generally applicable to other biomedical data, providing new algorithms for training models robust to bias for big data analysis and applications. Demo training code is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge