May D Wang
Autonomous Soft Tissue Retraction Using Demonstration-Guided Reinforcement Learning
Sep 02, 2023Abstract:In the context of surgery, robots can provide substantial assistance by performing small, repetitive tasks such as suturing, needle exchange, and tissue retraction, thereby enabling surgeons to concentrate on more complex aspects of the procedure. However, existing surgical task learning mainly pertains to rigid body interactions, whereas the advancement towards more sophisticated surgical robots necessitates the manipulation of soft bodies. Previous work focused on tissue phantoms for soft tissue task learning, which can be expensive and can be an entry barrier to research. Simulation environments present a safe and efficient way to learn surgical tasks before their application to actual tissue. In this study, we create a Robot Operating System (ROS)-compatible physics simulation environment with support for both rigid and soft body interactions within surgical tasks. Furthermore, we investigate the soft tissue interactions facilitated by the patient-side manipulator of the DaVinci surgical robot. Leveraging the pybullet physics engine, we simulate kinematics and establish anchor points to guide the robotic arm when manipulating soft tissue. Using demonstration-guided reinforcement learning (RL) algorithms, we investigate their performance in comparison to traditional reinforcement learning algorithms. Our in silico trials demonstrate a proof-of-concept for autonomous surgical soft tissue retraction. The results corroborate the feasibility of learning soft body manipulation through the application of reinforcement learning agents. This work lays the foundation for future research into the development and refinement of surgical robots capable of managing both rigid and soft tissue interactions. Code is available at https://github.com/amritpal-001/tissue_retract.
Improve Model Generalization and Robustness to Dataset Bias with Bias-regularized Learning and Domain-guided Augmentation
Nov 13, 2019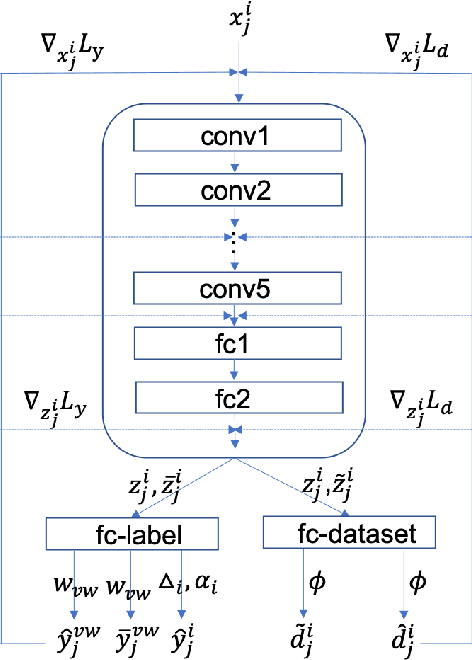
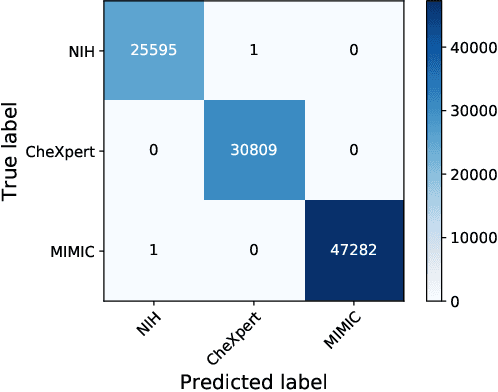
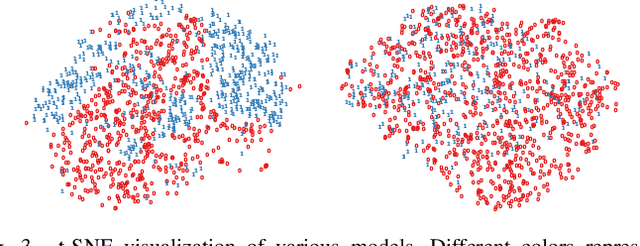
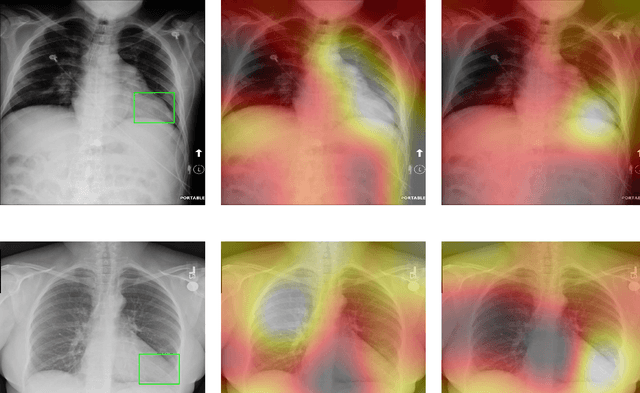
Abstract:Deep Learning has thrived on the emergence of biomedical big data. However, medical datasets acquired at different institutions have inherent bias caused by various confounding factors such as operation policies, machine protocols, treatment preference and etc. As the result, models trained on one dataset, regardless of volume, cannot be confidently utilized for the others. In this study, we investigated model robustness to dataset bias using three large-scale Chest X-ray datasets: first, we assessed the dataset bias using vanilla training baseline; second, we proposed a novel multi-source domain generalization model by (a) designing a new bias-regularized loss function; and (b) synthesizing new data for domain augmentation. We showed that our model significantly outperformed the baseline and other approaches on data from unseen domain in terms of accuracy and various bias measures, without retraining or finetuning. Our method is generally applicable to other biomedical data, providing new algorithms for training models robust to bias for big data analysis and applications. Demo training code is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge