Hedieh Sajedi
Unsupervised Anomaly Detection with an Enhanced Teacher for Student-Teacher Feature Pyramid Matching
Dec 20, 2025



Abstract:Anomaly detection or outlier is one of the challenging subjects in unsupervised learning . This paper is introduced a student-teacher framework for anomaly detection that its teacher network is enhanced for achieving high-performance metrics . For this purpose , we first pre-train the ResNet-18 network on the ImageNet and then fine-tune it on the MVTech-AD dataset . Experiment results on the image-level and pixel-level demonstrate that this idea has achieved better metrics than the previous methods . Our model , Enhanced Teacher for Student-Teacher Feature Pyramid (ET-STPM), achieved 0.971 mean accuracy on the image-level and 0.977 mean accuracy on the pixel-level for anomaly detection.
IoMT-based Automated Leukemia Classification using CNN and Higher Order Singular Value
Dec 18, 2025



Abstract:The Internet of Things (IoT) is a concept by which objects find identity and can communicate with each other in a network. One of the applications of the IoT is in the field of medicine, which is called the Internet of Medical Things (IoMT). Acute Lymphocytic Leukemia (ALL) is a type of cancer categorized as a hematic disease. It usually begins in the bone marrow due to the overproduction of immature White Blood Cells (WBCs or leukocytes). Since it has a high rate of spread to other body organs, it is a fatal disease if not diagnosed and treated early. Therefore, for identifying cancerous (ALL) cells in medical diagnostic laboratories, blood, as well as bone marrow smears, are taken by pathologists. However, manual examinations face limitations due to human error risk and time-consuming procedures. So, to tackle the mentioned issues, methods based on Artificial Intelligence (AI), capable of identifying cancer from non-cancer tissue, seem vital. Deep Neural Networks (DNNs) are the most efficient machine learning (ML) methods. These techniques employ multiple layers to extract higher-level features from the raw input. In this paper, a Convolutional Neural Network (CNN) is applied along with a new type of classifier, Higher Order Singular Value Decomposition (HOSVD), to categorize ALL and normal (healthy) cells from microscopic blood images. We employed the model on IoMT structure to identify leukemia quickly and safely. With the help of this new leukemia classification framework, patients and clinicians can have real-time communication. The model was implemented on the Acute Lymphoblastic Leukemia Image Database (ALL-IDB2) and achieved an average accuracy of %98.88 in the test step.
AI-Driven Relocation Tracking in Dynamic Kitchen Environments
Mar 03, 2025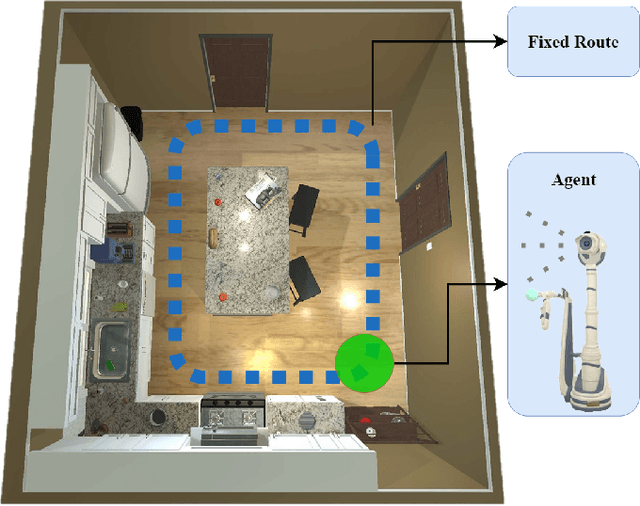

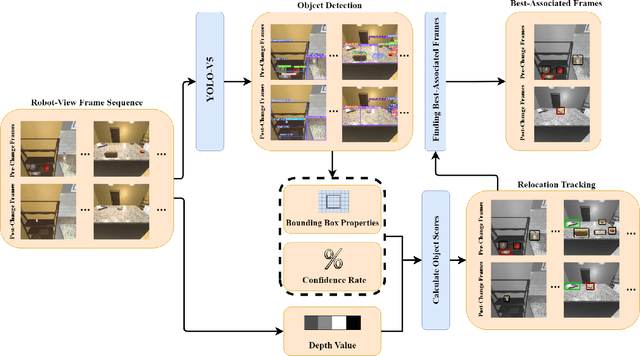
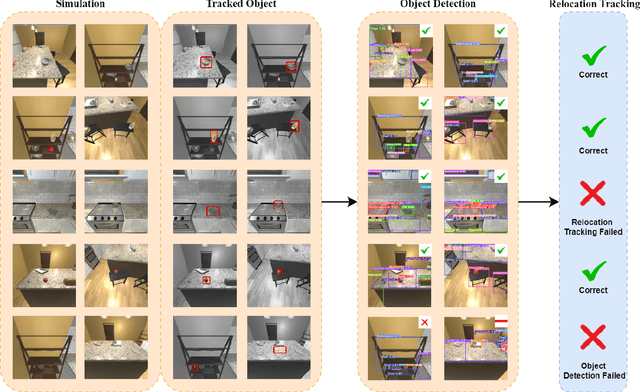
Abstract:As smart homes become more prevalent in daily life, the ability to understand dynamic environments is essential which is increasingly dependent on AI systems. This study focuses on developing an intelligent algorithm which can navigate a robot through a kitchen, recognizing objects, and tracking their relocation. The kitchen was chosen as the testing ground due to its dynamic nature as objects are frequently moved, rearranged and replaced. Various techniques, such as SLAM feature-based tracking and deep learning-based object detection (e.g., Faster R-CNN), are commonly used for object tracking. Additionally, methods such as optical flow analysis and 3D reconstruction have also been used to track the relocation of objects. These approaches often face challenges when it comes to problems such as lighting variations and partial occlusions, where parts of the object are hidden in some frames but visible in others. The proposed method in this study leverages the YOLOv5 architecture, initialized with pre-trained weights and subsequently fine-tuned on a custom dataset. A novel method was developed, introducing a frame-scoring algorithm which calculates a score for each object based on its location and features within all frames. This scoring approach helps to identify changes by determining the best-associated frame for each object and comparing the results in each scene, overcoming limitations seen in other methods while maintaining simplicity in design. The experimental results demonstrate an accuracy of 97.72%, a precision of 95.83% and a recall of 96.84% for this algorithm, which highlights the efficacy of the model in detecting spatial changes.
Segmentation of Coronary Artery Stenosis in X-ray Angiography using Mamba Models
Dec 03, 2024Abstract:Coronary artery disease stands as one of the primary contributors to global mortality rates. The automated identification of coronary artery stenosis from X-ray images plays a critical role in the diagnostic process for coronary heart disease. This task is challenging due to the complex structure of coronary arteries, intrinsic noise in X-ray images, and the fact that stenotic coronary arteries appear narrow and blurred in X-ray angiographies. This study employs five different variants of the Mamba-based model and one variant of the Swin Transformer-based model, primarily based on the U-Net architecture, for the localization of stenosis in Coronary artery disease. Our best results showed an F1 score of 68.79% for the U-Mamba BOT model, representing an 11.8% improvement over the semi-supervised approach.
Brain Age Estimation with a Greedy Dual-Stream Model for Limited Datasets
Jul 05, 2024



Abstract:Brain age estimation involves predicting the biological age of individuals from their brain images, which offers valuable insights into the aging process and the progression of neurodegenerative diseases. Conducting large-scale datasets for medical image analysis is a challenging and time-consuming task. Existing approaches mostly depend on large datasets, which are hard to come by and expensive. These approaches also require sophisticated, resource-intensive models with a large number of parameters, necessitating a considerable amount of processing power. As a result, there is a vital need to develop innovative methods that can achieve robust performance with limited datasets and efficient use of computational resources. This paper proposes a novel slice-based dual-stream method called GDSM (Greedy Dual-Stream Model) for brain age estimation. This method addresses the limitations of large dataset requirements and computational resource intensiveness. The proposed method incorporates local and global aspects of the brain, thereby refining the focus on specific target regions. The approach employs four backbones to predict ages based on local and global features, complemented by a final model for age correction. Our method demonstrates a Mean Absolute Error (MAE) of 3.25 years on the test set of IBID, which only contains 289 subjects. To demonstrate the robustness of our approach for any small dataset, we analyzed the proposed method with the IXI dataset and achieved an MAE of 4.18 years on the test set of IXI. By leveraging dual-stream and greedy strategies, this approach achieves efficiency and robust performance, making it comparable with other state-of-the-art methods. The code for the GDSM model is available at https://github.com/iman2693/GDSM.
Predict joint angle of body parts based on sequence pattern recognition
May 27, 2024Abstract:The way organs are positioned and moved in the workplace can cause pain and physical harm. Therefore, ergonomists use ergonomic risk assessments based on visual observation of the workplace, or review pictures and videos taken in the workplace. Sometimes the workers in the photos are not in perfect condition. Some parts of the workers' bodies may not be in the camera's field of view, could be obscured by objects, or by self-occlusion, this is the main problem in 2D human posture recognition. It is difficult to predict the position of body parts when they are not visible in the image, and geometric mathematical methods are not entirely suitable for this purpose. Therefore, we created a dataset with artificial images of a 3D human model, specifically for painful postures, and real human photos from different viewpoints. Each image we captured was based on a predefined joint angle for each 3D model or human model. We created various images, including images where some body parts are not visible. Nevertheless, the joint angle is estimated beforehand, so we could study the case by converting the input images into the sequence of joint connections between predefined body parts and extracting the desired joint angle with a convolutional neural network. In the end, we obtained root mean square error (RMSE) of 12.89 and mean absolute error (MAE) of 4.7 on the test dataset.
Hand bone age estimation using divide and conquer strategy and lightweight convolutional neural networks
May 23, 2024Abstract:Estimating the Bone Age of children is very important for diagnosing growth defects, and related diseases, and estimating the final height that children reach after maturity. For this reason, it is widely used in different countries. Traditional methods for estimating bone age are performed by comparing atlas images and radiographic images of the left hand, which is time-consuming and error-prone. To estimate bone age using deep neural network models, a lot of research has been done, our effort has been to improve the accuracy and speed of this process by using the introduced approach. After creating and analyzing our initial model, we focused on preprocessing and made the inputs smaller, and increased their quality. we selected small regions of hand radiographs and estimated the age of the bone only according to these regions. by doing this we improved bone age estimation accuracy even further than what was achieved in related works, without increasing the required computational resource. We reached a Mean Absolute Error (MAE) of 3.90 months in the range of 0-20 years and an MAE of 3.84 months in the range of 1-18 years on the RSNA test set.
Gallbladder Cancer Detection in Ultrasound Images based on YOLO and Faster R-CNN
Apr 23, 2024



Abstract:Medical image analysis is a significant application of artificial intelligence for disease diagnosis. A crucial step in this process is the identification of regions of interest within the images. This task can be automated using object detection algorithms. YOLO and Faster R-CNN are renowned for such algorithms, each with its own strengths and weaknesses. This study aims to explore the advantages of both techniques to select more accurate bounding boxes for gallbladder detection from ultrasound images, thereby enhancing gallbladder cancer classification. A fusion method that leverages the benefits of both techniques is presented in this study. The proposed method demonstrated superior classification performance, with an accuracy of 92.62%, compared to the individual use of Faster R-CNN and YOLOv8, which yielded accuracies of 90.16% and 82.79%, respectively.
* Published in 2024 10th International Conference on Artificial Intelligence and Robotics (QICAR)
A survey on automated detection and classification of acute leukemia and WBCs in microscopic blood cells
Mar 10, 2023Abstract:Leukemia (blood cancer) is an unusual spread of White Blood Cells or Leukocytes (WBCs) in the bone marrow and blood. Pathologists can diagnose leukemia by looking at a person's blood sample under a microscope. They identify and categorize leukemia by counting various blood cells and morphological features. This technique is time-consuming for the prediction of leukemia. The pathologist's professional skills and experiences may be affecting this procedure, too. In computer vision, traditional machine learning and deep learning techniques are practical roadmaps that increase the accuracy and speed in diagnosing and classifying medical images such as microscopic blood cells. This paper provides a comprehensive analysis of the detection and classification of acute leukemia and WBCs in the microscopic blood cells. First, we have divided the previous works into six categories based on the output of the models. Then, we describe various steps of detection and classification of acute leukemia and WBCs, including Data Augmentation, Preprocessing, Segmentation, Feature Extraction, Feature Selection (Reduction), Classification, and focus on classification step in the methods. Finally, we divide automated detection and classification of acute leukemia and WBCs into three categories, including traditional, Deep Neural Network (DNN), and mixture (traditional and DNN) methods based on the type of classifier in the classification step and analyze them. The results of this study show that in the diagnosis and classification of acute leukemia and WBCs, the Support Vector Machine (SVM) classifier in traditional machine learning models and Convolutional Neural Network (CNN) classifier in deep learning models have widely employed. The performance metrics of the models that use these classifiers compared to the others model are higher.
Convolutional Neural Network-Based Image Watermarking using Discrete Wavelet Transform
Oct 08, 2022



Abstract:As the Internet becomes more popular, digital images are used and transferred more frequently. Although this phenomenon facilitates easy access to information, it also creates security concerns and violates intellectual property rights by allowing illegal use, copying, and digital content theft. Using watermarks (WMs) in digital images is one of the most common ways to maintain security. Watermarking is proving and declaring ownership of an image by adding a digital watermark to the original image. Watermarks can be either text or an image placed overtly or covertly in an image and are expected to be challenging to remove. This paper proposes a combination of convolutional neural networks (CNNs) and wavelet transforms to obtain a watermarking network for embedding and extracting watermarks. The network is independent of the host image resolution, can accept all kinds of watermarks, and has only 11 CNN layers while keeping performance. Two terms measure performance; the similarity between the extracted watermark and the original one and the similarity between the host image and the watermarked one.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge