Hang Lou
PCF-GAN: generating sequential data via the characteristic function of measures on the path space
May 21, 2023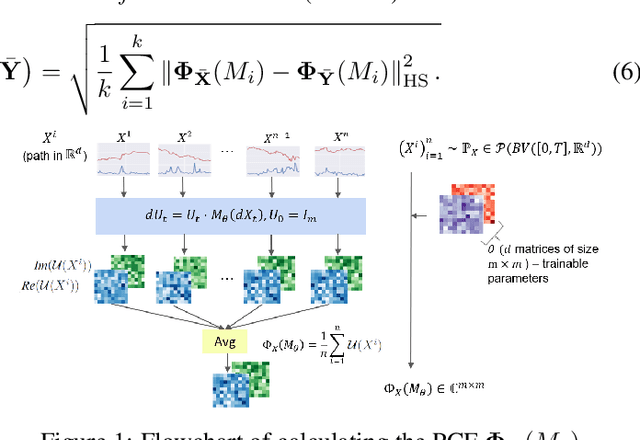

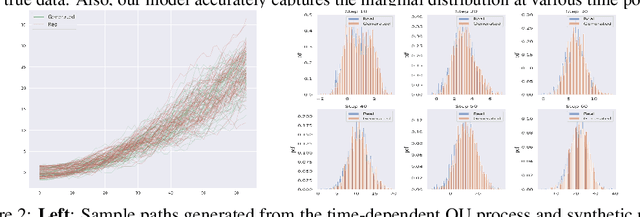
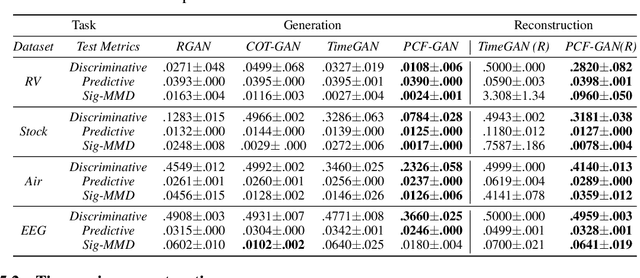
Abstract:Generating high-fidelity time series data using generative adversarial networks (GANs) remains a challenging task, as it is difficult to capture the temporal dependence of joint probability distributions induced by time-series data. Towards this goal, a key step is the development of an effective discriminator to distinguish between time series distributions. We propose the so-called PCF-GAN, a novel GAN that incorporates the path characteristic function (PCF) as the principled representation of time series distribution into the discriminator to enhance its generative performance. On the one hand, we establish theoretical foundations of the PCF distance by proving its characteristicity, boundedness, differentiability with respect to generator parameters, and weak continuity, which ensure the stability and feasibility of training the PCF-GAN. On the other hand, we design efficient initialisation and optimisation schemes for PCFs to strengthen the discriminative power and accelerate training efficiency. To further boost the capabilities of complex time series generation, we integrate the auto-encoder structure via sequential embedding into the PCF-GAN, which provides additional reconstruction functionality. Extensive numerical experiments on various datasets demonstrate the consistently superior performance of PCF-GAN over state-of-the-art baselines, in both generation and reconstruction quality. Code is available at https://github.com/DeepIntoStreams/PCF-GAN.
Beating the Best: Improving on AlphaFold2 at Protein Structure Prediction
Jan 23, 2023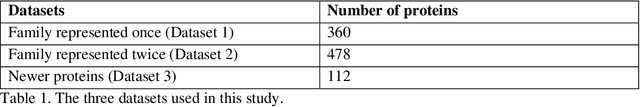
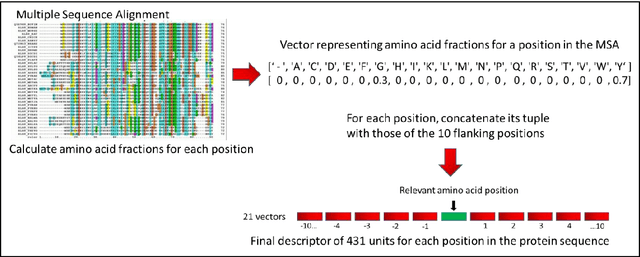
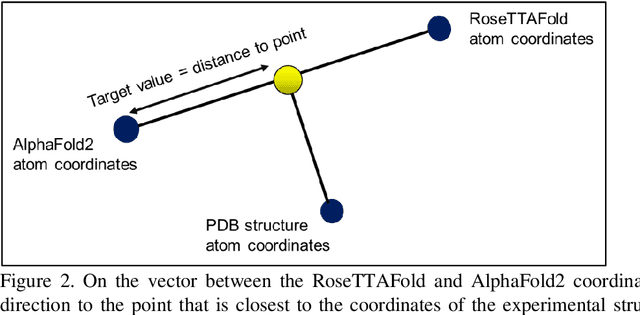
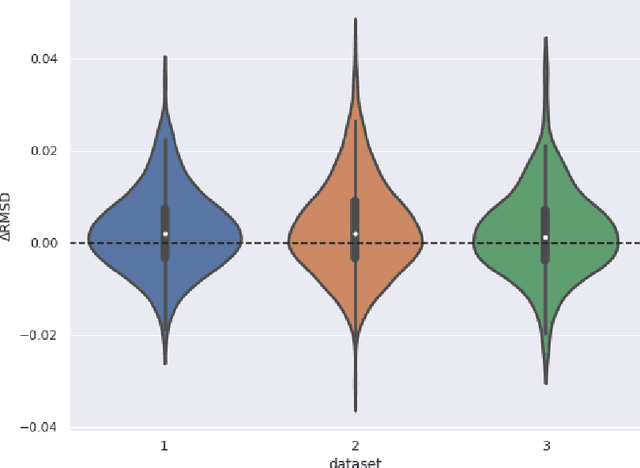
Abstract:The goal of Protein Structure Prediction (PSP) problem is to predict a protein's 3D structure (confirmation) from its amino acid sequence. The problem has been a 'holy grail' of science since the Noble prize-winning work of Anfinsen demonstrated that protein conformation was determined by sequence. A recent and important step towards this goal was the development of AlphaFold2, currently the best PSP method. AlphaFold2 is probably the highest profile application of AI to science. Both AlphaFold2 and RoseTTAFold (another impressive PSP method) have been published and placed in the public domain (code & models). Stacking is a form of ensemble machine learning ML in which multiple baseline models are first learnt, then a meta-model is learnt using the outputs of the baseline level model to form a model that outperforms the base models. Stacking has been successful in many applications. We developed the ARStack PSP method by stacking AlphaFold2 and RoseTTAFold. ARStack significantly outperforms AlphaFold2. We rigorously demonstrate this using two sets of non-homologous proteins, and a test set of protein structures published after that of AlphaFold2 and RoseTTAFold. As more high quality prediction methods are published it is likely that ensemble methods will increasingly outperform any single method.
Path Development Network with Finite-dimensional Lie Group Representation
Apr 02, 2022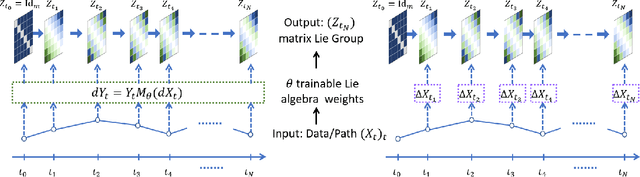
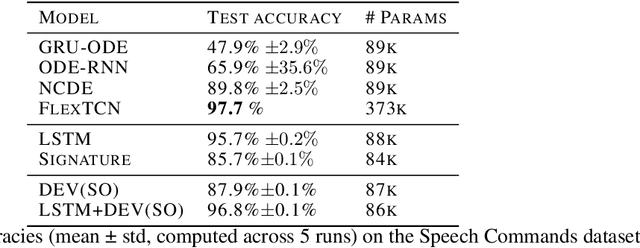
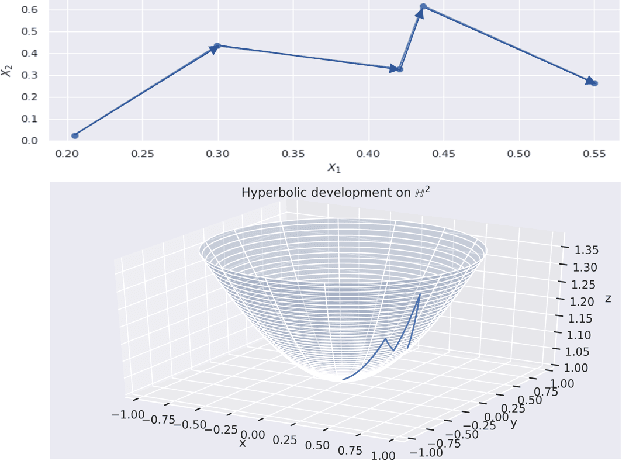
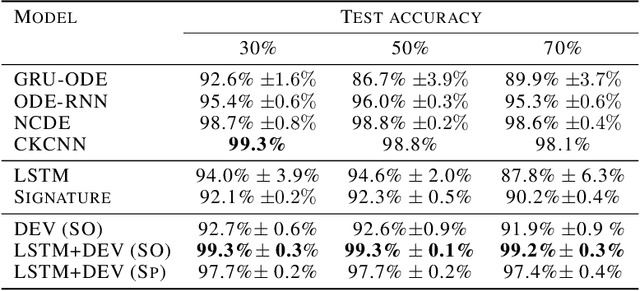
Abstract:The path signature, a mathematically principled and universal feature of sequential data, leads to a performance boost of deep learning-based models in various sequential data tasks as a complimentary feature. However, it suffers from the curse of dimensionality when the path dimension is high. To tackle this problem, we propose a novel, trainable path development layer, which exploits representations of sequential data with the help of finite-dimensional matrix Lie groups. We also design the backpropagation algorithm of the development layer via an optimisation method on manifolds known as trivialisation. Numerical experiments demonstrate that the path development consistently and significantly outperforms, in terms of accuracy and dimensionality, signature features on several empirical datasets. Moreover, stacking the LSTM with the development layer with a suitable matrix Lie group is empirically proven to alleviate the gradient issues of LSTMs and the resulting hybrid model achieves the state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge