Beating the Best: Improving on AlphaFold2 at Protein Structure Prediction
Paper and Code
Jan 23, 2023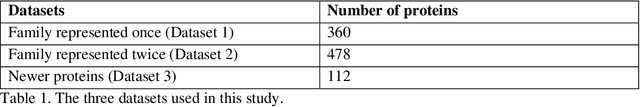
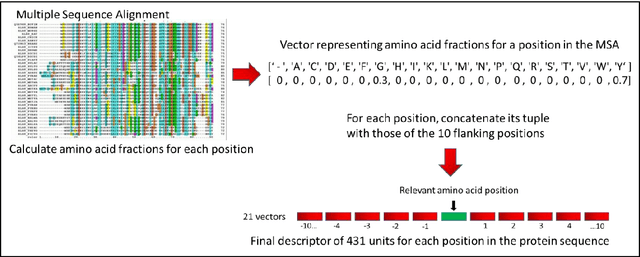
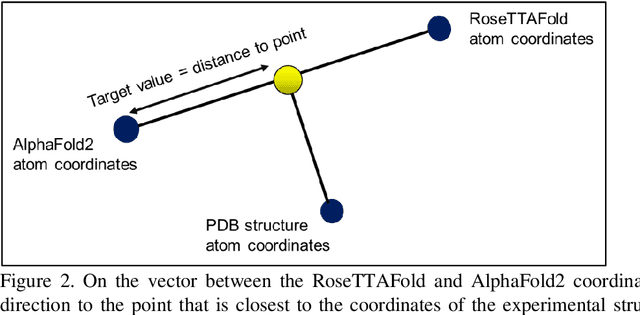
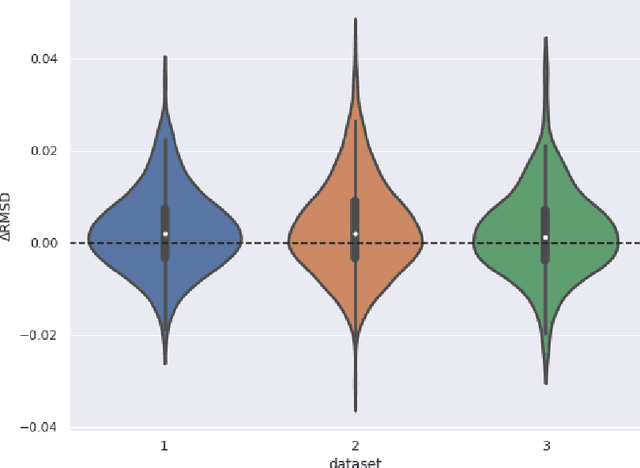
The goal of Protein Structure Prediction (PSP) problem is to predict a protein's 3D structure (confirmation) from its amino acid sequence. The problem has been a 'holy grail' of science since the Noble prize-winning work of Anfinsen demonstrated that protein conformation was determined by sequence. A recent and important step towards this goal was the development of AlphaFold2, currently the best PSP method. AlphaFold2 is probably the highest profile application of AI to science. Both AlphaFold2 and RoseTTAFold (another impressive PSP method) have been published and placed in the public domain (code & models). Stacking is a form of ensemble machine learning ML in which multiple baseline models are first learnt, then a meta-model is learnt using the outputs of the baseline level model to form a model that outperforms the base models. Stacking has been successful in many applications. We developed the ARStack PSP method by stacking AlphaFold2 and RoseTTAFold. ARStack significantly outperforms AlphaFold2. We rigorously demonstrate this using two sets of non-homologous proteins, and a test set of protein structures published after that of AlphaFold2 and RoseTTAFold. As more high quality prediction methods are published it is likely that ensemble methods will increasingly outperform any single method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge