Hakime Öztürk
Exploring Chemical Space using Natural Language Processing Methodologies for Drug Discovery
Feb 10, 2020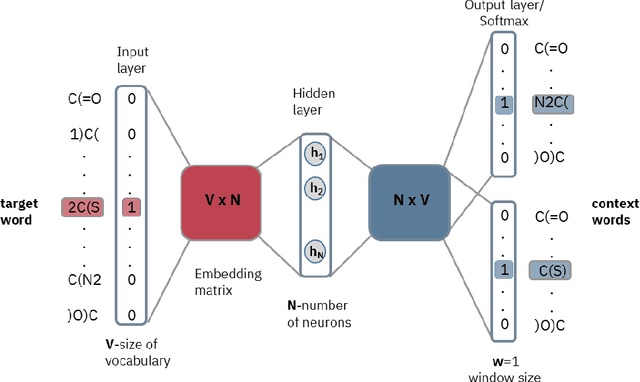
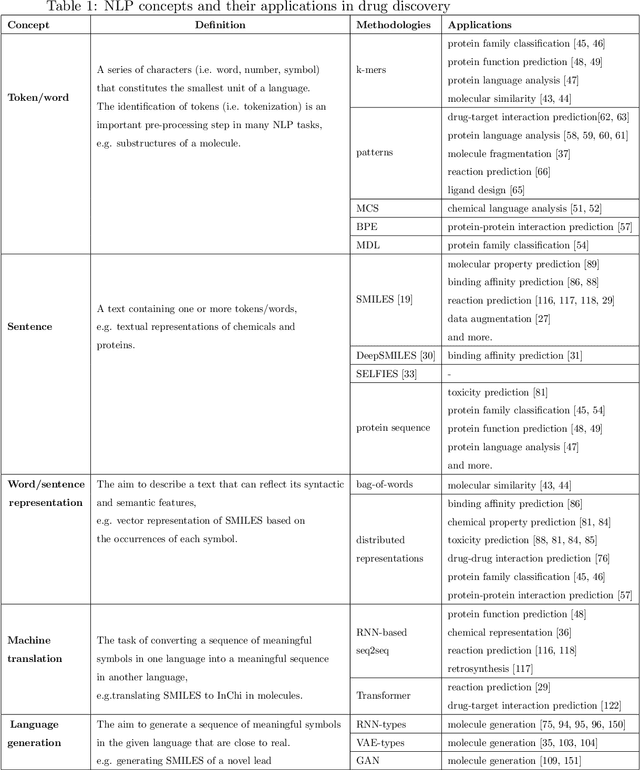
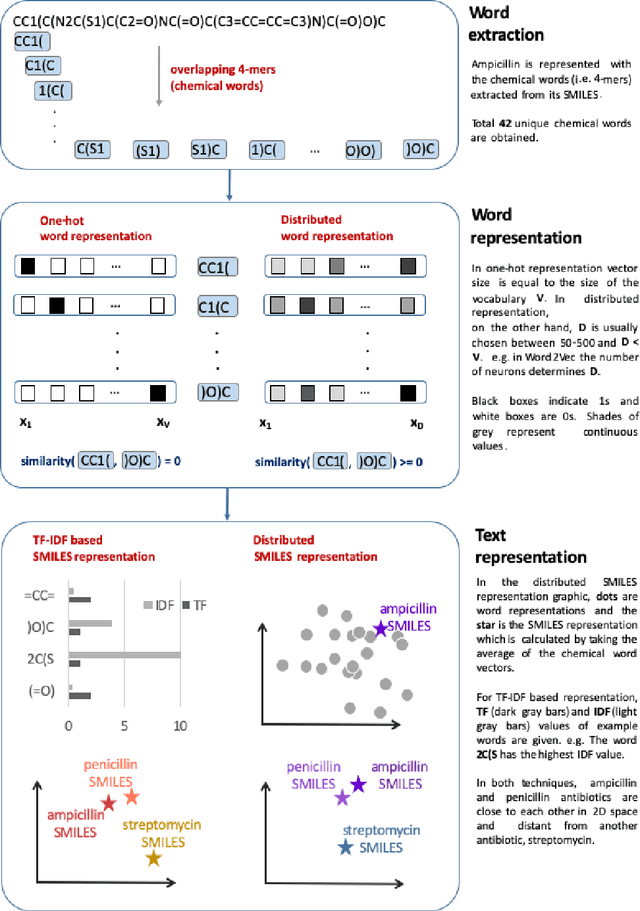
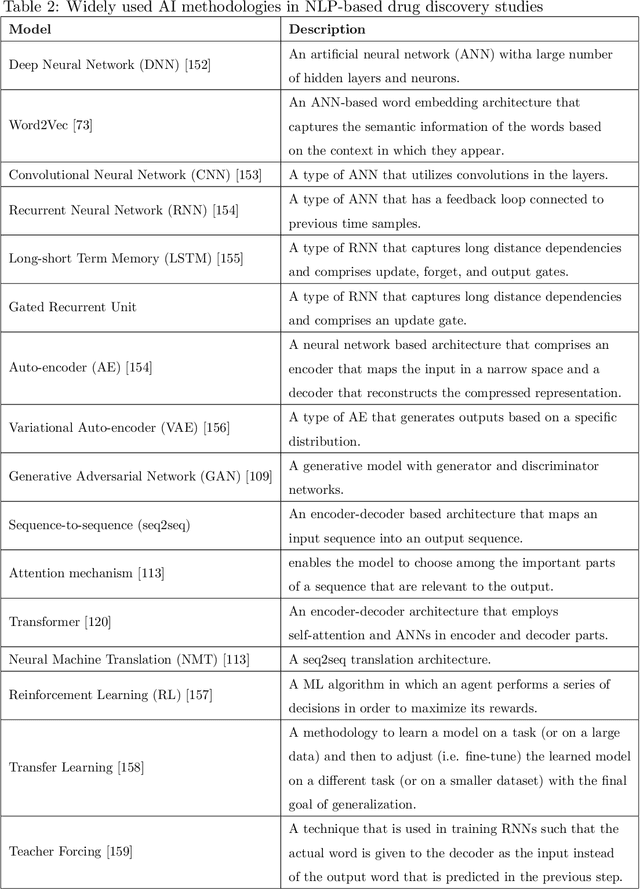
Abstract:Text-based representations of chemicals and proteins can be thought of as unstructured languages codified by humans to describe domain-specific knowledge. Advances in natural language processing (NLP) methodologies in the processing of spoken languages accelerated the application of NLP to elucidate hidden knowledge in textual representations of these biochemical entities and then use it to construct models to predict molecular properties or to design novel molecules. This review outlines the impact made by these advances on drug discovery and aims to further the dialogue between medicinal chemists and computer scientists.
WideDTA: prediction of drug-target binding affinity
Feb 04, 2019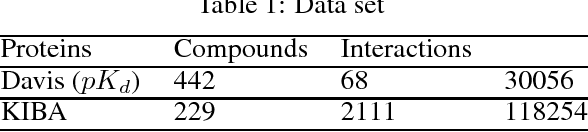
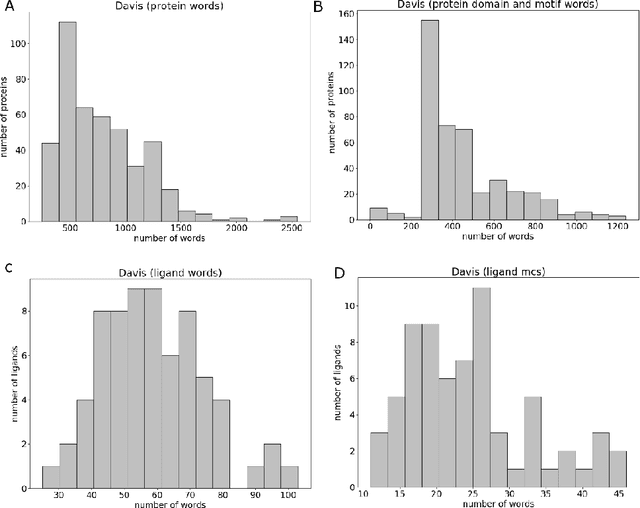
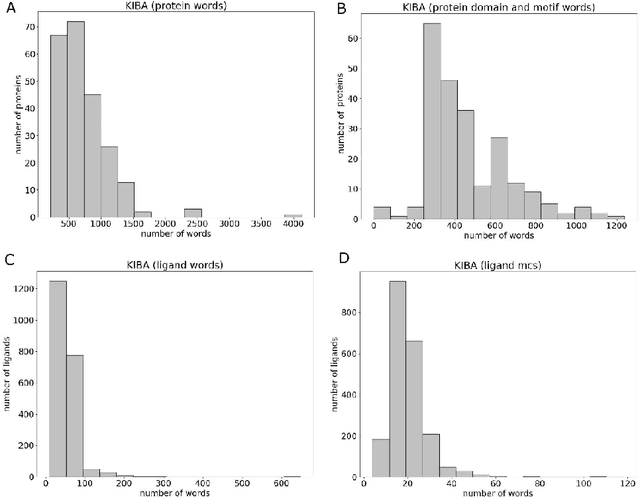
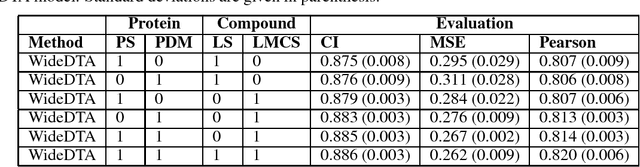
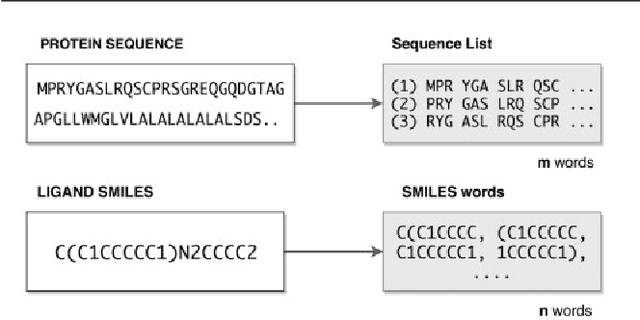
Abstract:Motivation: Prediction of the interaction affinity between proteins and compounds is a major challenge in the drug discovery process. WideDTA is a deep-learning based prediction model that employs chemical and biological textual sequence information to predict binding affinity. Results: WideDTA uses four text-based information sources, namely the protein sequence, ligand SMILES, protein domains and motifs, and maximum common substructure words to predict binding affinity. WideDTA outperformed one of the state of the art deep learning methods for drug-target binding affinity prediction, DeepDTA on the KIBA dataset with a statistical significance. This indicates that the word-based sequence representation adapted by WideDTA is a promising alternative to the character-based sequence representation approach in deep learning models for binding affinity prediction, such as the one used in DeepDTA. In addition, the results showed that, given the protein sequence and ligand SMILES, the inclusion of protein domain and motif information as well as ligand maximum common substructure words do not provide additional useful information for the deep learning model. Interestingly, however, using only domain and motif information to represent proteins achieved similar performance to using the full protein sequence, suggesting that important binding relevant information is contained within the protein motifs and domains.
A chemical language based approach for protein - ligand interaction prediction
Nov 02, 2018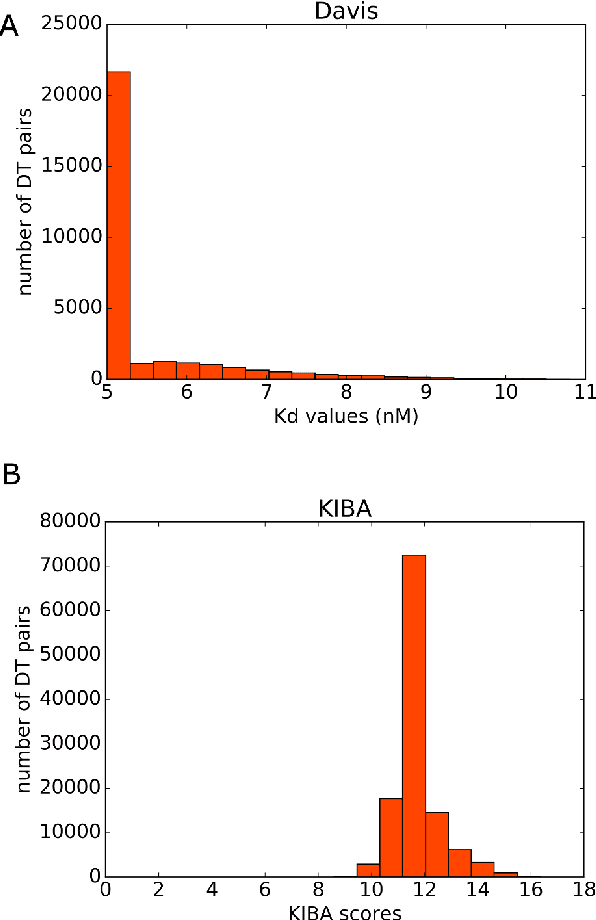
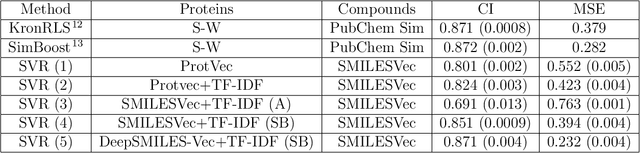
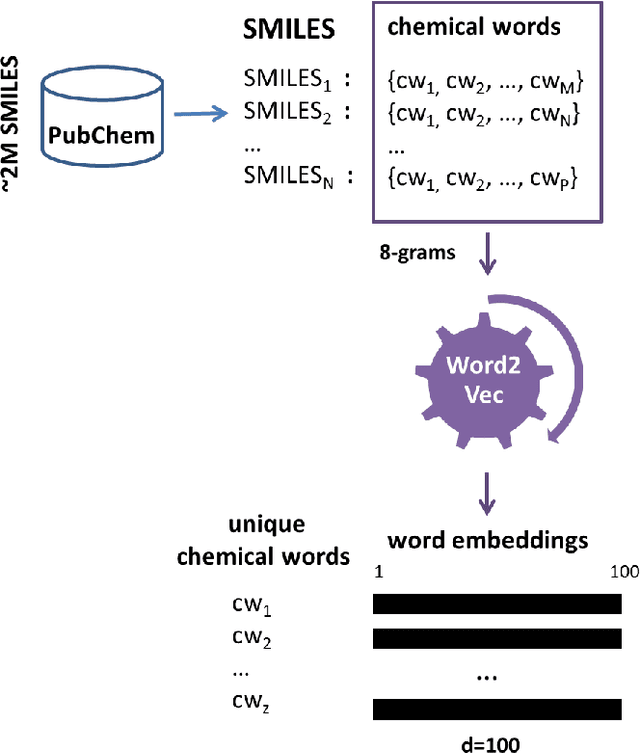
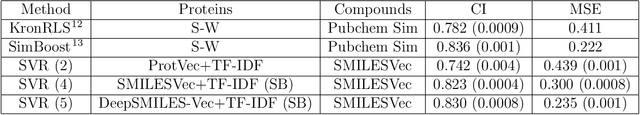
Abstract:Identification of high affinity drug-target interactions (DTI) is a major research question in drug discovery. In this study, we propose a novel methodology to predict drug-target binding affinity using only ligand SMILES information. We represent proteins using the word-embeddings of the SMILES representations of their strong binding ligands. Each SMILES is represented in the form of a set of chemical words and a protein is described by the set of chemical words with the highest Term Frequency- Inverse Document Frequency (TF-IDF) value. We then utilize the Support Vector Regression (SVR) algorithm to predict protein - drug binding affinities in the Davis and KIBA Kinase datasets. We also compared the performance of SMILES representation with the recently proposed DeepSMILES representation and found that using DeepSMILES yields better performance in the prediction task. Using only SMILESVec, which is a strictly string based representation of the proteins based on their interacting ligands, we were able to predict drug-target binding affinity as well as or better than the KronRLS or SimBoost models that utilize protein sequence.
DeepDTA: Deep Drug-Target Binding Affinity Prediction
Jun 05, 2018
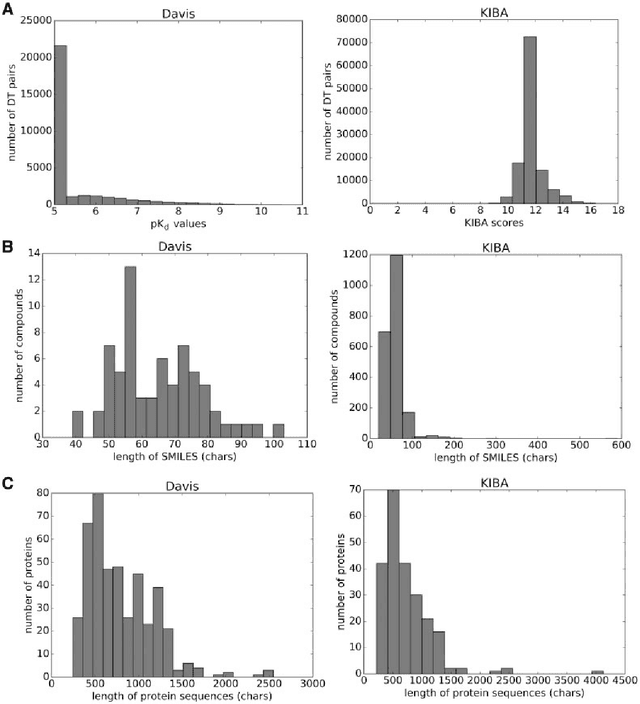
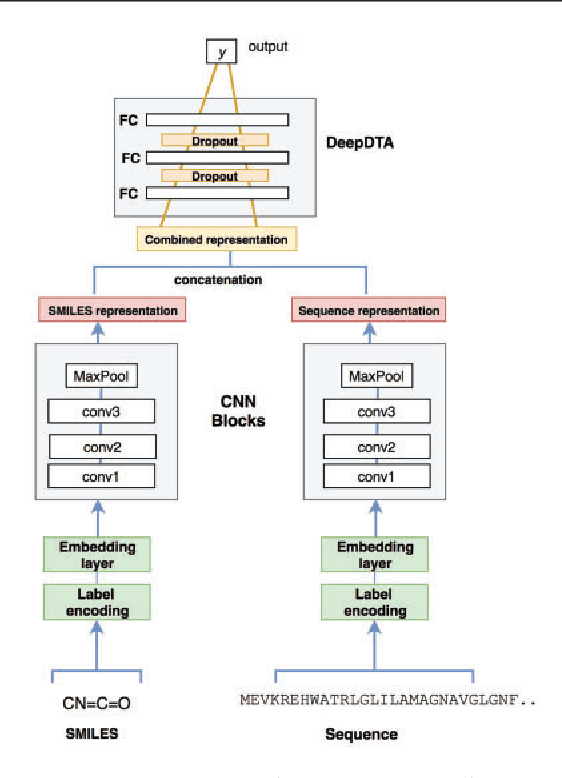

Abstract:The identification of novel drug-target (DT) interactions is a substantial part of the drug discovery process. Most of the computational methods that have been proposed to predict DT interactions have focused on binary classification, where the goal is to determine whether a DT pair interacts or not. However, protein-ligand interactions assume a continuum of binding strength values, also called binding affinity and predicting this value still remains a challenge. The increase in the affinity data available in DT knowledge-bases allows the use of advanced learning techniques such as deep learning architectures in the prediction of binding affinities. In this study, we propose a deep-learning based model that uses only sequence information of both targets and drugs to predict DT interaction binding affinities. The few studies that focus on DT binding affinity prediction use either 3D structures of protein-ligand complexes or 2D features of compounds. One novel approach used in this work is the modeling of protein sequences and compound 1D representations with convolutional neural networks (CNNs). The results show that the proposed deep learning based model that uses the 1D representations of targets and drugs is an effective approach for drug target binding affinity prediction. The model in which high-level representations of a drug and a target are constructed via CNNs achieved the best Concordance Index (CI) performance in one of our larger benchmark data sets, outperforming the KronRLS algorithm and SimBoost, a state-of-the-art method for DT binding affinity prediction.
A novel methodology on distributed representations of proteins using their interacting ligands
Jan 30, 2018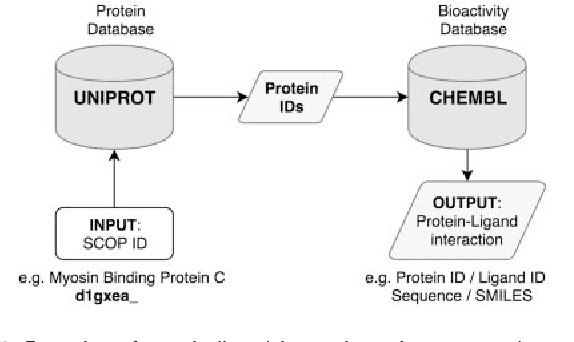

Abstract:The effective representation of proteins is a crucial task that directly affects the performance of many bioinformatics problems. Related proteins usually bind to similar ligands. Chemical characteristics of ligands are known to capture the functional and mechanistic properties of proteins suggesting that a ligand based approach can be utilized in protein representation. In this study, we propose SMILESVec, a SMILES-based method to represent ligands and a novel method to compute similarity of proteins by describing them based on their ligands. The proteins are defined utilizing the word-embeddings of the SMILES strings of their ligands. The performance of the proposed protein description method is evaluated in protein clustering task using TransClust and MCL algorithms. Two other protein representation methods that utilize protein sequence, BLAST and ProtVec, and two compound fingerprint based protein representation methods are compared. We showed that ligand-based protein representation, which uses only SMILES strings of the ligands that proteins bind to, performs as well as protein-sequence based representation methods in protein clustering. The results suggest that ligand-based protein description can be an alternative to the traditional sequence or structure based representation of proteins and this novel approach can be applied to different bioinformatics problems such as prediction of new protein-ligand interactions and protein function annotation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge