H Juliette T Unwin
Deep learning and MCMC with aggVAE for shifting administrative boundaries: mapping malaria prevalence in Kenya
May 31, 2023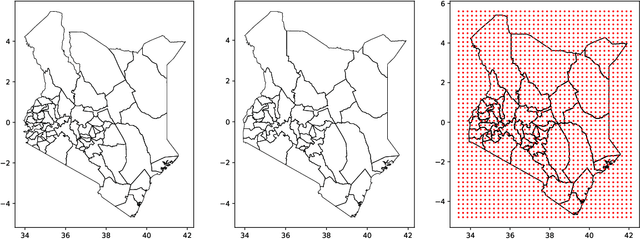
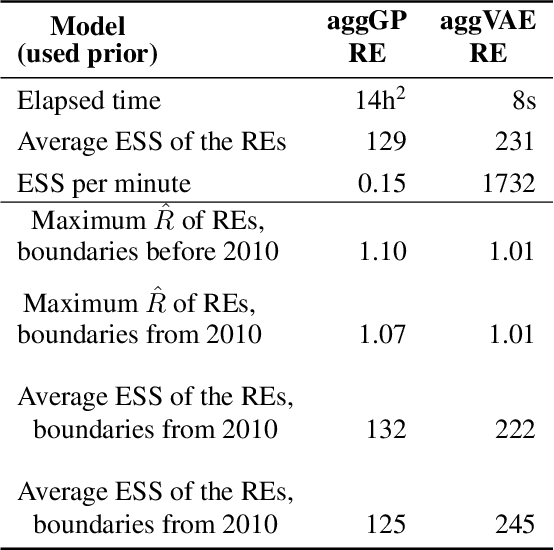
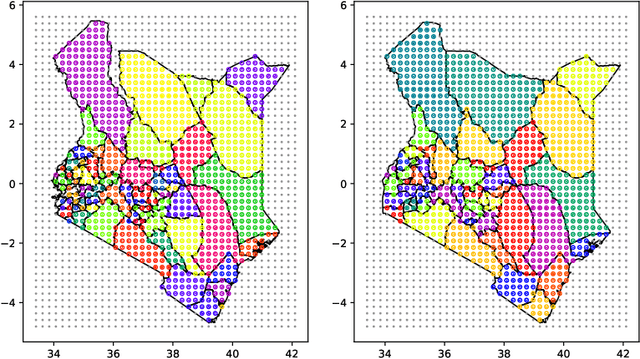

Abstract:Model-based disease mapping remains a fundamental policy-informing tool in public health and disease surveillance with hierarchical Bayesian models being the current state-of-the-art approach. When working with areal data, e.g. aggregates at the administrative unit level such as district or province, routinely used models rely on the adjacency structure of areal units to account for spatial correlations. The goal of disease surveillance systems is to track disease outcomes over time, but this provides challenging in situations of crises, such as political changes, leading to changes of administrative boundaries. Kenya is an example of such country. Moreover, adjacency-based approach ignores the continuous nature of spatial processes and cannot solve the change-of-support problem, i.e. when administrative boundaries change. We present a novel, practical, and easy to implement solution relying on a methodology combining deep generative modelling and fully Bayesian inference. We build on the recent work of PriorVAE able to encode spatial priors over small areas with variational autoencoders, to map malaria prevalence in Kenya. We solve the change-of-support problem arising from Kenya changing its district boundaries in 2010. We draw realisations of the Gaussian Process (GP) prior over a fine artificial spatial grid representing continuous space and then aggregate these realisations to the level of administrative boundaries. The aggregated values are then encoded using the PriorVAE technique. The trained priors (aggVAE) are then used at the inference stage instead of the GP priors within a Markov chain Monte Carlo (MCMC) scheme. We demonstrate that it is possible to use the flexible and appropriate model for areal data based on aggregation of continuous priors, and that inference is orders of magnitude faster when using aggVAE than combining the original GP priors and the aggregation step.
A comparison of short-term probabilistic forecasts for the incidence of COVID-19 using mechanistic and statistical time series models
May 01, 2023Abstract:Short-term forecasts of infectious disease spread are a critical component in risk evaluation and public health decision making. While different models for short-term forecasting have been developed, open questions about their relative performance remain. Here, we compare short-term probabilistic forecasts of popular mechanistic models based on the renewal equation with forecasts of statistical time series models. Our empirical comparison is based on data of the daily incidence of COVID-19 across six large US states over the first pandemic year. We find that, on average, probabilistic forecasts from statistical time series models are overall at least as accurate as forecasts from mechanistic models. Moreover, statistical time series models better capture volatility. Our findings suggest that domain knowledge, which is integrated into mechanistic models by making assumptions about disease dynamics, does not improve short-term forecasts of disease incidence. We note, however, that forecasting is often only one of many objectives and thus mechanistic models remain important, for example, to model the impact of vaccines or the emergence of new variants.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge