György Cserey
VReID-XFD: Video-based Person Re-identification at Extreme Far Distance Challenge Results
Jan 04, 2026Abstract:Person re-identification (ReID) across aerial and ground views at extreme far distances introduces a distinct operating regime where severe resolution degradation, extreme viewpoint changes, unstable motion cues, and clothing variation jointly undermine the appearance-based assumptions of existing ReID systems. To study this regime, we introduce VReID-XFD, a video-based benchmark and community challenge for extreme far-distance (XFD) aerial-to-ground person re-identification. VReID-XFD is derived from the DetReIDX dataset and comprises 371 identities, 11,288 tracklets, and 11.75 million frames, captured across altitudes from 5.8 m to 120 m, viewing angles from oblique (30 degrees) to nadir (90 degrees), and horizontal distances up to 120 m. The benchmark supports aerial-to-aerial, aerial-to-ground, and ground-to-aerial evaluation under strict identity-disjoint splits, with rich physical metadata. The VReID-XFD-25 Challenge attracted 10 teams with hundreds of submissions. Systematic analysis reveals monotonic performance degradation with altitude and distance, a universal disadvantage of nadir views, and a trade-off between peak performance and robustness. Even the best-performing SAS-PReID method achieves only 43.93 percent mAP in the aerial-to-ground setting. The dataset, annotations, and official evaluation protocols are publicly available at https://www.it.ubi.pt/DetReIDX/ .
Deep Anomaly Generation: An Image Translation Approach of Synthesizing Abnormal Banded Chromosome Images
Sep 20, 2021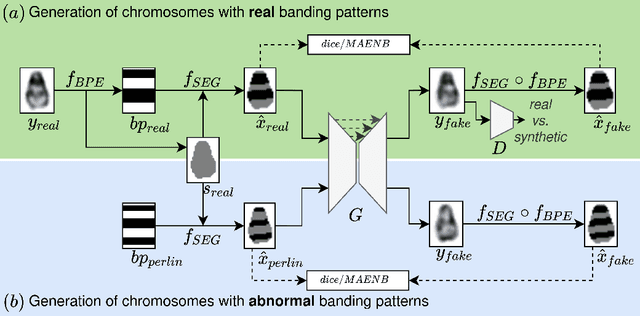
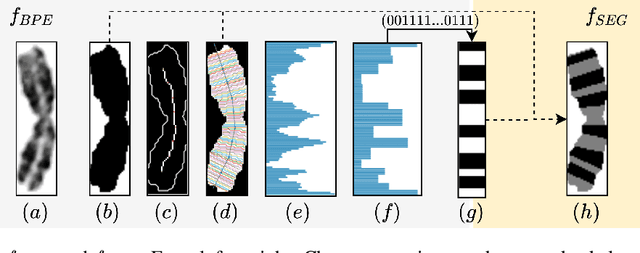
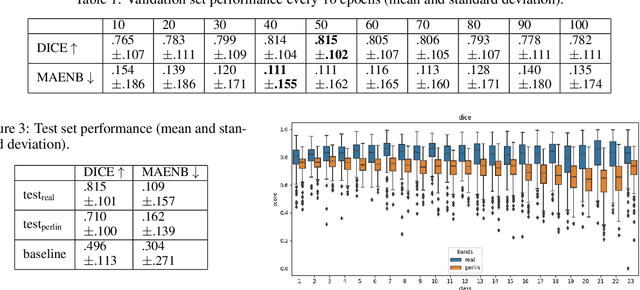
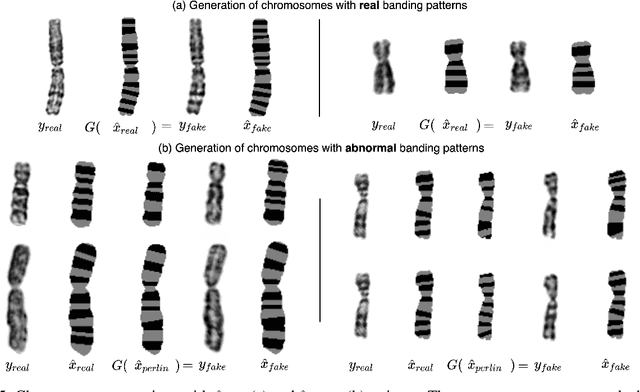
Abstract:Advances in deep-learning-based pipelines have led to breakthroughs in a variety of microscopy image diagnostics. However, a sufficiently big training data set is usually difficult to obtain due to high annotation costs. In the case of banded chromosome images, the creation of big enough libraries is difficult for multiple pathologies due to the rarity of certain genetic disorders. Generative Adversarial Networks (GANs) have proven to be effective in generating synthetic images and extending training data sets. In our work, we implement a conditional adversarial network that allows generation of realistic single chromosome images following user-defined banding patterns. To this end, an image-to-image translation approach based on self-generated 2D chromosome segmentation label maps is used. Our validation shows promising results when synthesizing chromosomes with seen as well as unseen banding patterns. We believe that this approach can be exploited for data augmentation of chromosome data sets with structural abnormalities. Therefore, the proposed method could help to tackle medical image analysis problems such as data simulation, segmentation, detection, or classification in the field of cytogenetics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge