Giltae Song
CAST: Cluster-Aware Self-Training for Tabular Data
Oct 10, 2023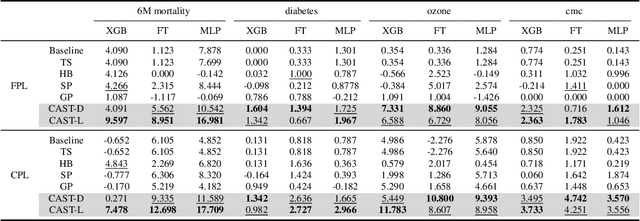
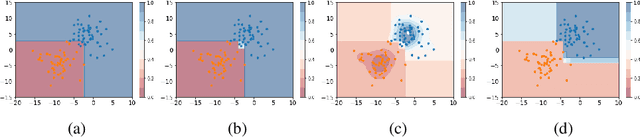
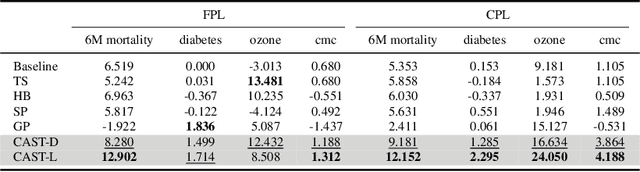
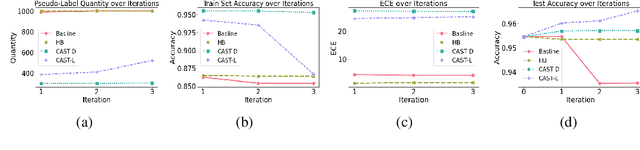
Abstract:Self-training has gained attraction because of its simplicity and versatility, yet it is vulnerable to noisy pseudo-labels. Several studies have proposed successful approaches to tackle this issue, but they have diminished the advantages of self-training because they require specific modifications in self-training algorithms or model architectures. Furthermore, most of them are incompatible with gradient boosting decision trees, which dominate the tabular domain. To address this, we revisit the cluster assumption, which states that data samples that are close to each other tend to belong to the same class. Inspired by the assumption, we propose Cluster-Aware Self-Training (CAST) for tabular data. CAST is a simple and universally adaptable approach for enhancing existing self-training algorithms without significant modifications. Concretely, our method regularizes the confidence of the classifier, which represents the value of the pseudo-label, forcing the pseudo-labels in low-density regions to have lower confidence by leveraging prior knowledge for each class within the training data. Extensive empirical evaluations on up to 20 real-world datasets confirm not only the superior performance of CAST but also its robustness in various setups in self-training contexts.
Revisiting Self-Training with Regularized Pseudo-Labeling for Tabular Data
Mar 13, 2023Abstract:Recent progress in semi- and self-supervised learning has caused a rift in the long-held belief about the need for an enormous amount of labeled data for machine learning and the irrelevancy of unlabeled data. Although it has been successful in various data, there is no dominant semi- and self-supervised learning method that can be generalized for tabular data (i.e. most of the existing methods require appropriate tabular datasets and architectures). In this paper, we revisit self-training which can be applied to any kind of algorithm including the most widely used architecture, gradient boosting decision tree, and introduce curriculum pseudo-labeling (a state-of-the-art pseudo-labeling technique in image) for a tabular domain. Furthermore, existing pseudo-labeling techniques do not assure the cluster assumption when computing confidence scores of pseudo-labels generated from unlabeled data. To overcome this issue, we propose a novel pseudo-labeling approach that regularizes the confidence scores based on the likelihoods of the pseudo-labels so that more reliable pseudo-labels which lie in high density regions can be obtained. We exhaustively validate the superiority of our approaches using various models and tabular datasets.
Bayesian History Reconstruction of Complex Human Gene Clusters on a Phylogeny
Jun 15, 2009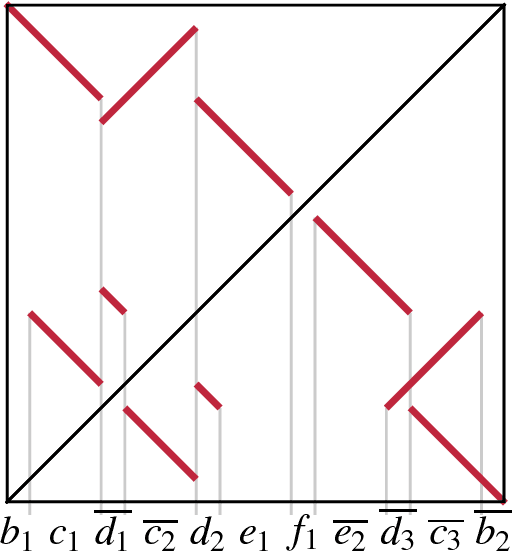
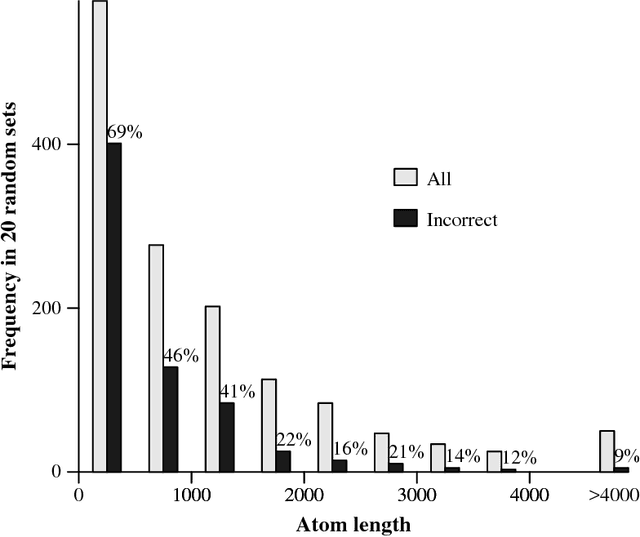
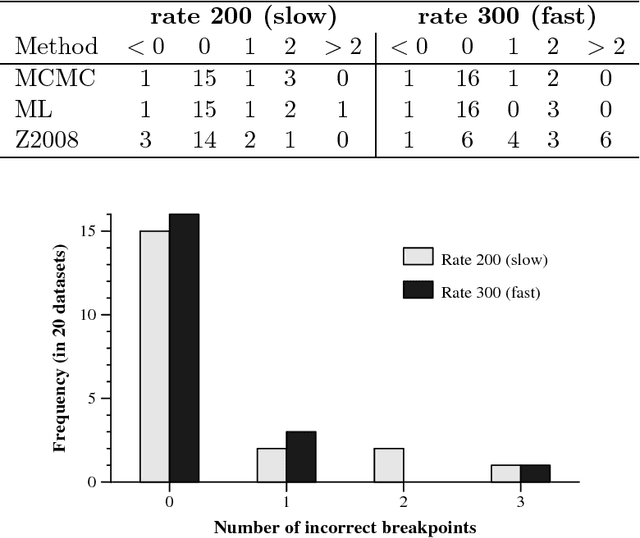
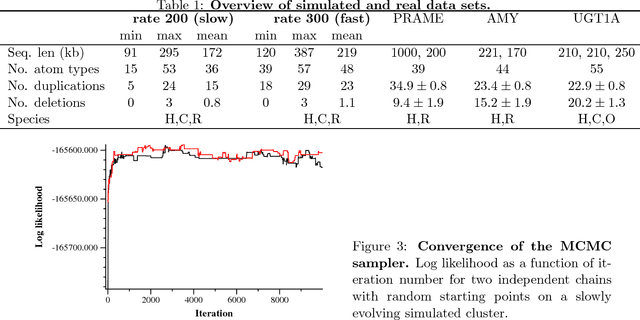
Abstract:Clusters of genes that have evolved by repeated segmental duplication present difficult challenges throughout genomic analysis, from sequence assembly to functional analysis. Improved understanding of these clusters is of utmost importance, since they have been shown to be the source of evolutionary innovation, and have been linked to multiple diseases, including HIV and a variety of cancers. Previously, Zhang et al. (2008) developed an algorithm for reconstructing parsimonious evolutionary histories of such gene clusters, using only human genomic sequence data. In this paper, we propose a probabilistic model for the evolution of gene clusters on a phylogeny, and an MCMC algorithm for reconstruction of duplication histories from genomic sequences in multiple species. Several projects are underway to obtain high quality BAC-based assemblies of duplicated clusters in multiple species, and we anticipate that our method will be useful in analyzing these valuable new data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge