Adam Siepel
Bayesian History Reconstruction of Complex Human Gene Clusters on a Phylogeny
Jun 15, 2009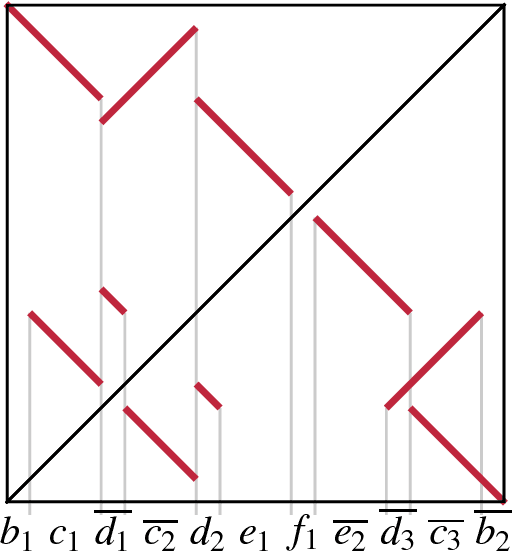
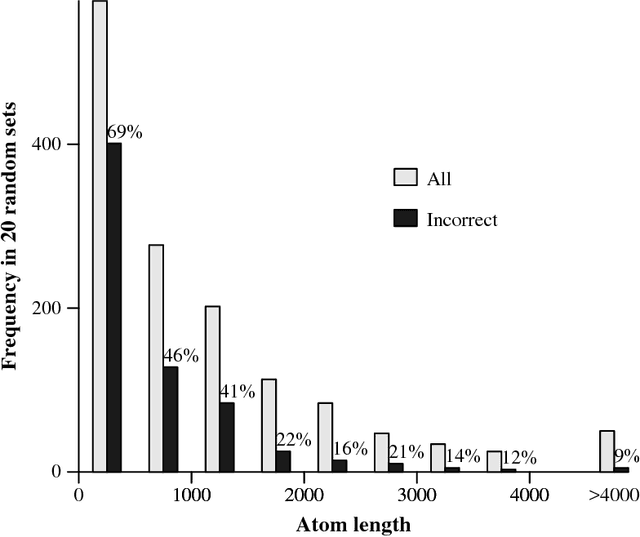
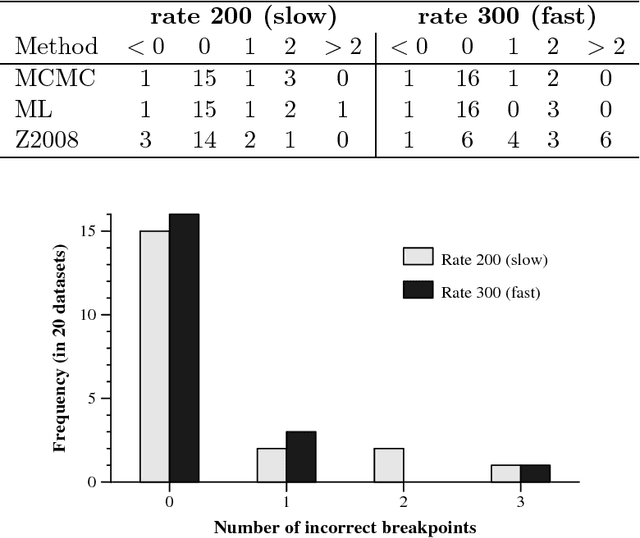
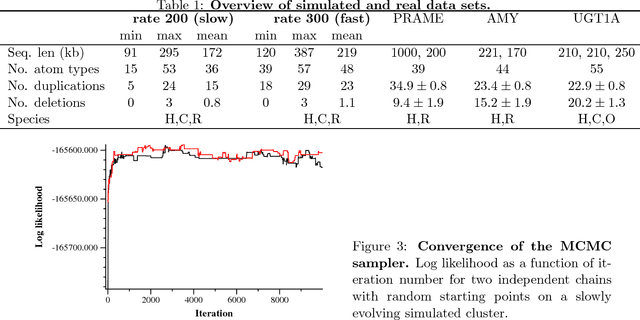
Abstract:Clusters of genes that have evolved by repeated segmental duplication present difficult challenges throughout genomic analysis, from sequence assembly to functional analysis. Improved understanding of these clusters is of utmost importance, since they have been shown to be the source of evolutionary innovation, and have been linked to multiple diseases, including HIV and a variety of cancers. Previously, Zhang et al. (2008) developed an algorithm for reconstructing parsimonious evolutionary histories of such gene clusters, using only human genomic sequence data. In this paper, we propose a probabilistic model for the evolution of gene clusters on a phylogeny, and an MCMC algorithm for reconstruction of duplication histories from genomic sequences in multiple species. Several projects are underway to obtain high quality BAC-based assemblies of duplicated clusters in multiple species, and we anticipate that our method will be useful in analyzing these valuable new data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge