George Nehmetallah
Virtual organelle self-coding for fluorescence imaging via adversarial learning
Sep 10, 2019



Abstract:Fluorescence microscopy plays a vital role in understanding the subcellular structures of living cells. However, it requires considerable effort in sample preparation related to chemical fixation, staining, cost, and time. To reduce those factors, we present a virtual fluorescence staining method based on deep neural networks (VirFluoNet) to transform fluorescence images of molecular labels into other molecular fluorescence labels in the same field-of-view. To achieve this goal, we develop and train a conditional generative adversarial network (cGAN) to perform digital fluorescence imaging demonstrated on human osteosarcoma U2OS cell fluorescence images captured under Cell Painting staining protocol. A detailed comparative analysis is also conducted on the performance of the cGAN network between predicting fluorescence channels based on phase contrast or based on another fluorescence channel using human breast cancer MDA-MB-231 cell line as a test case. In addition, we implement a deep learning model to perform autofocusing on another human U2OS fluorescence dataset as a preprocessing step to defocus an out-focus channel in U2OS dataset. A quantitative index of image prediction error is introduced based on signal pixel-wise spatial and intensity differences with ground truth to evaluate the performance of prediction to high-complex and throughput fluorescence. This index provides a rational way to perform image segmentation on error signals and to understand the likelihood of mis-interpreting biology from the predicted image. In total, these findings contribute to the utility of deep learning image regression for fluorescence microscopy datasets of biological cells, balanced against savings of cost, time, and experimental effort. Furthermore, the approach introduced here holds promise for modeling the internal relationships between organelles and biomolecules within living cells.
Deep learning approach to Fourier ptychographic microscopy
Jul 30, 2018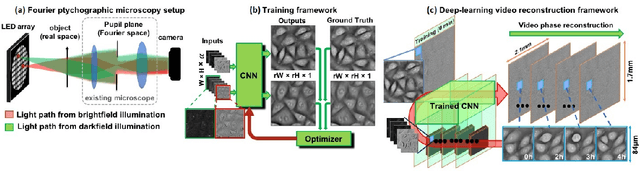
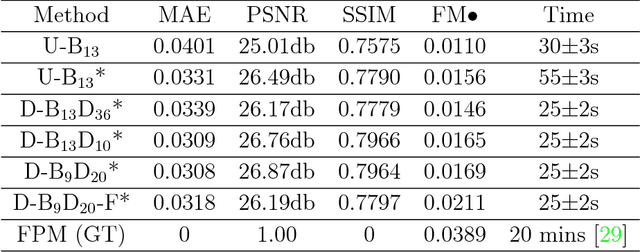
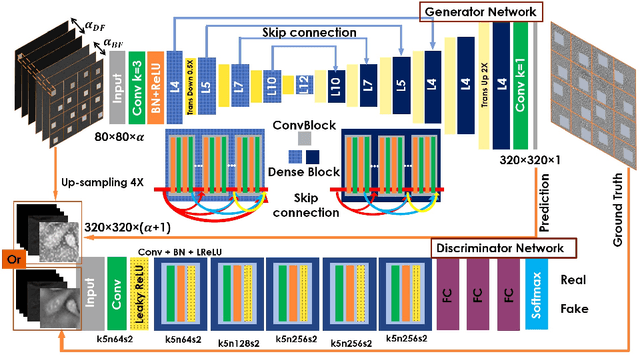
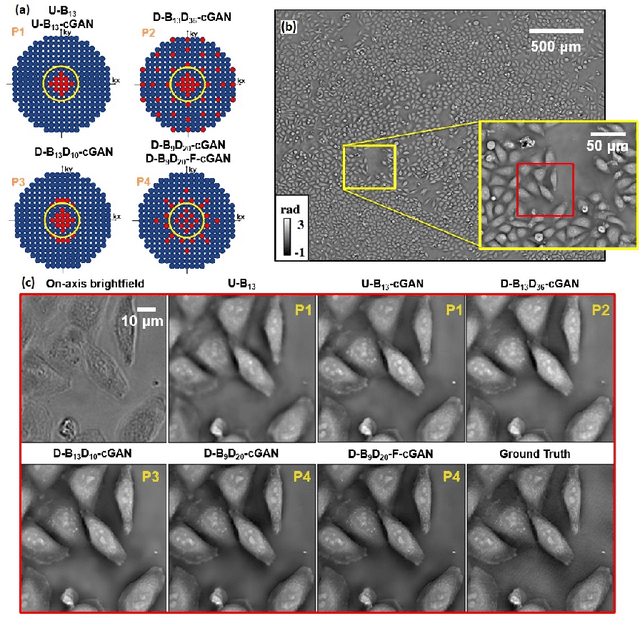
Abstract:Convolutional neural networks (CNNs) have gained tremendous success in solving complex inverse problems. The aim of this work is to develop a novel CNN framework to reconstruct video sequence of dynamic live cells captured using a computational microscopy technique, Fourier ptychographic microscopy (FPM). The unique feature of the FPM is its capability to reconstruct images with both wide field-of-view (FOV) and high resolution, i.e. a large space-bandwidth-product (SBP), by taking a series of low resolution intensity images. For live cell imaging, a single FPM frame contains thousands of cell samples with different morphological features. Our idea is to fully exploit the statistical information provided by this large spatial ensemble so as to make predictions in a sequential measurement, without using any additional temporal dataset. Specifically, we show that it is possible to reconstruct high-SBP dynamic cell videos by a CNN trained only on the first FPM dataset captured at the beginning of a time-series experiment. Our CNN approach reconstructs a 12800X10800 pixels phase image using only ~25 seconds, a 50X speedup compared to the model-based FPM algorithm. In addition, the CNN further reduces the required number of images in each time frame by ~6X. Overall, this significantly improves the imaging throughput by reducing both the acquisition and computational times. The proposed CNN is based on the conditional generative adversarial network (cGAN) framework. Additionally, we also exploit transfer learning so that our pre-trained CNN can be further optimized to image other cell types. Our technique demonstrates a promising deep learning approach to continuously monitor large live-cell populations over an extended time and gather useful spatial and temporal information with sub-cellular resolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge