George M. Coghill
A Survey on Physarum Polycephalum Intelligent Foraging Behaviour and Bio-Inspired Applications
Mar 07, 2021



Abstract:In recent years, research on Physarum polycephalum has become more popular after Nakagaki et al. (2000) performed their famous experiment showing that Physarum was able to find the shortest route through a maze. Subsequent researches have confirmed the ability of Physarum-inspired algorithms to solve a wide range of NP-hard problems. This review will through light on recent Physarum polycephalum biological aspects, mathematical models, and Physarum bio-inspired algorithms and their applications. Further, we have presented our new model to simulate Physarum in competition, where multiple Physarum interact with each other and with their environments. The bio-inspired Physarum in competition algorithms proved to have great potentials for future research.
A Novel Genetic Algorithm with Hierarchical Evaluation Strategy for Hyperparameter Optimisation of Graph Neural Networks
Jan 26, 2021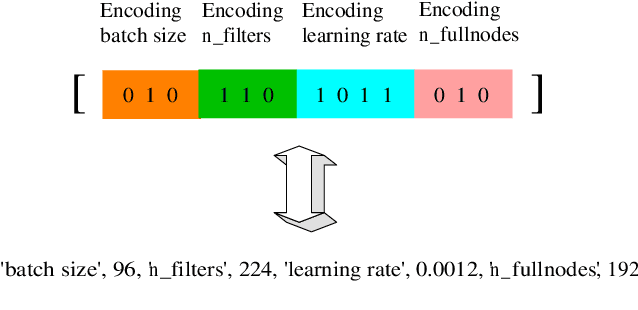
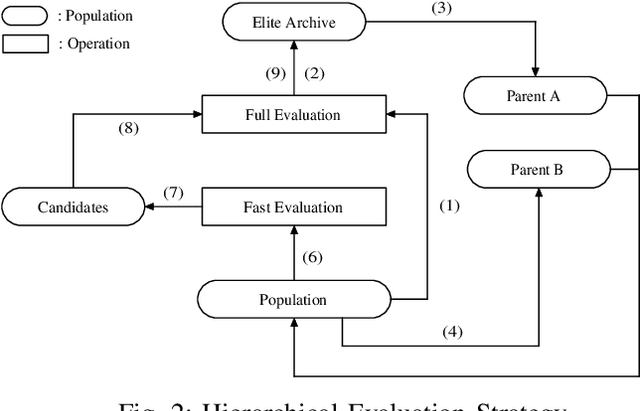
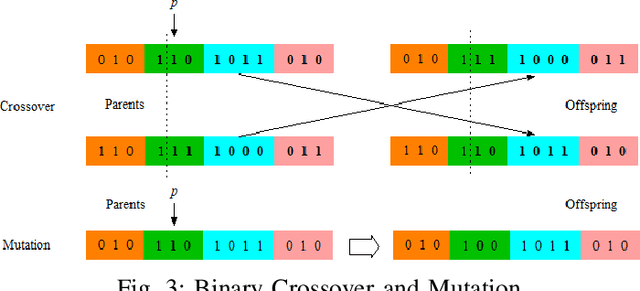
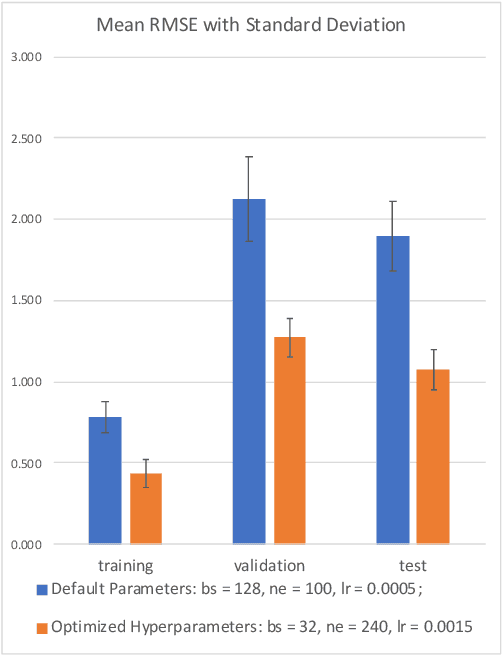
Abstract:Graph representation of structured data can facilitate the extraction of stereoscopic features, and it has demonstrated excellent ability when working with deep learning systems, the so-called Graph Neural Networks (GNNs). Choosing a promising architecture for constructing GNNs can be transferred to a hyperparameter optimisation problem, a very challenging task due to the size of the underlying search space and high computational cost for evaluating candidate GNNs. To address this issue, this research presents a novel genetic algorithm with a hierarchical evaluation strategy (HESGA), which combines the full evaluation of GNNs with a fast evaluation approach. By using full evaluation, a GNN is represented by a set of hyperparameter values and trained on a specified dataset, and root mean square error (RMSE) will be used to measure the quality of the GNN represented by the set of hyperparameter values (for regression problems). While in the proposed fast evaluation process, the training will be interrupted at an early stage, the difference of RMSE values between the starting and interrupted epochs will be used as a fast score, which implies the potential of the GNN being considered. To coordinate both types of evaluations, the proposed hierarchical strategy uses the fast evaluation in a lower level for recommending candidates to a higher level, where the full evaluation will act as a final assessor to maintain a group of elite individuals. To validate the effectiveness of HESGA, we apply it to optimise two types of deep graph neural networks. The experimental results on three benchmark datasets demonstrate its advantages compared to Bayesian hyperparameter optimization.
ImmuNeCS: Neural Committee Search by an Artificial Immune System
Nov 19, 2019



Abstract:Current Neural Architecture Search techniques can suffer from a few shortcomings, including high computational cost, excessive bias from the search space, conceptual complexity or uncertain empirical benefits over random search. In this paper, we present ImmuNeCS, an attempt at addressing these issues with a method that offers a simple, flexible, and efficient way of building deep learning models automatically, and we demonstrate its effectiveness in the context of convolutional neural networks. Instead of searching for the 1-best architecture for a given task, we focus on building a population of neural networks that are then ensembled into a neural network committee, an approach we dub 'Neural Committee Search'. To ensure sufficient performance from the committee, our search algorithm is based on an artificial immune system that balances individual performance with population diversity. This allows us to stop the search when accuracy starts to plateau, and to bridge the performance gap through ensembling. In order to justify our method, we first verify that the chosen search space exhibits the locality property. To further improve efficiency, we also combine partial evaluation, weight inheritance, and progressive search. First, experiments are run to verify the validity of these techniques. Then, preliminary experimental results on two popular computer vision benchmarks show that our method consistently outperforms random search and yields promising results within reasonable GPU budgets. An additional experiment also shows that ImmuNeCS's solutions transfer effectively to a more difficult task, where they achieve results comparable to a direct search on the new task. We believe these findings can open the way for new, accessible alternatives to traditional NAS.
Qualitative System Identification from Imperfect Data
Oct 31, 2011



Abstract:Experience in the physical sciences suggests that the only realistic means of understanding complex systems is through the use of mathematical models. Typically, this has come to mean the identification of quantitative models expressed as differential equations. Quantitative modelling works best when the structure of the model (i.e., the form of the equations) is known; and the primary concern is one of estimating the values of the parameters in the model. For complex biological systems, the model-structure is rarely known and the modeler has to deal with both model-identification and parameter-estimation. In this paper we are concerned with providing automated assistance to the first of these problems. Specifically, we examine the identification by machine of the structural relationships between experimentally observed variables. These relationship will be expressed in the form of qualitative abstractions of a quantitative model. Such qualitative models may not only provide clues to the precise quantitative model, but also assist in understanding the essence of that model. Our position in this paper is that background knowledge incorporating system modelling principles can be used to constrain effectively the set of good qualitative models. Utilising the model-identification framework provided by Inductive Logic Programming (ILP) we present empirical support for this position using a series of increasingly complex artificial datasets. The results are obtained with qualitative and quantitative data subject to varying amounts of noise and different degrees of sparsity. The results also point to the presence of a set of qualitative states, which we term kernel subsets, that may be necessary for a qualitative model-learner to learn correct models. We demonstrate scalability of the method to biological system modelling by identification of the glycolysis metabolic pathway from data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge