Gennady Roshchupkin
Biomedical Imaging Group Rotterdam, Department of Radiology & Nuclear Medicine, Erasmus MC - University Medical Center Rotterdam, Rotterdam, the Netherlands, Department of Epidemiology, Erasmus MC - University Medical Center Rotterdam, Rotterdam, the Netherlands
MRI-based and metabolomics-based age scores act synergetically for mortality prediction shown by multi-cohort federated learning
Sep 02, 2024



Abstract:Biological age scores are an emerging tool to characterize aging by estimating chronological age based on physiological biomarkers. Various scores have shown associations with aging-related outcomes. This study assessed the relation between an age score based on brain MRI images (BrainAge) and an age score based on metabolomic biomarkers (MetaboAge). We trained a federated deep learning model to estimate BrainAge in three cohorts. The federated BrainAge model yielded significantly lower error for age prediction across the cohorts than locally trained models. Harmonizing the age interval between cohorts further improved BrainAge accuracy. Subsequently, we compared BrainAge with MetaboAge using federated association and survival analyses. The results showed a small association between BrainAge and MetaboAge as well as a higher predictive value for the time to mortality of both scores combined than for the individual scores. Hence, our study suggests that both aging scores capture different aspects of the aging process.
Projection-wise Disentangling for Fair and Interpretable Representation Learning: Application to 3D Facial Shape Analysis
Jun 28, 2021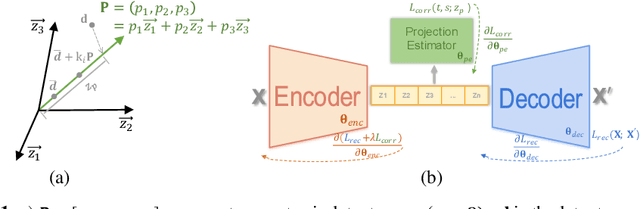
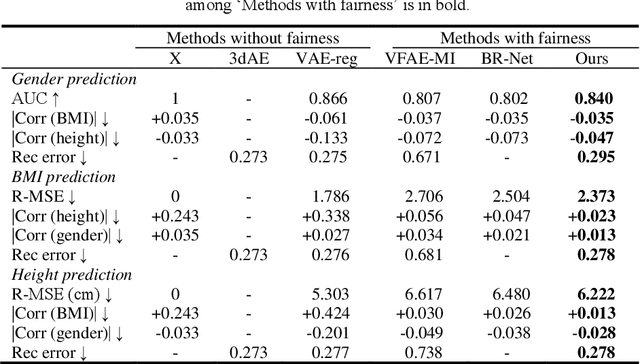
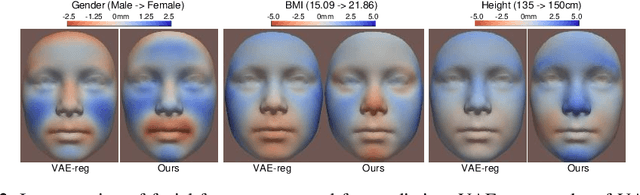
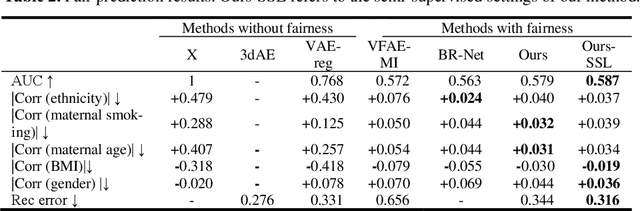
Abstract:Confounding bias is a crucial problem when applying machine learning to practice, especially in clinical practice. We consider the problem of learning representations independent to multiple biases. In literature, this is mostly solved by purging the bias information from learned representations. We however expect this strategy to harm the diversity of information in the representation, and thus limiting its prospective usage (e.g., interpretation). Therefore, we propose to mitigate the bias while keeping almost all information in the latent representations, which enables us to observe and interpret them as well. To achieve this, we project latent features onto a learned vector direction, and enforce the independence between biases and projected features rather than all learned features. To interpret the mapping between projected features and input data, we propose projection-wise disentangling: a sampling and reconstruction along the learned vector direction. The proposed method was evaluated on the analysis of 3D facial shape and patient characteristics (N=5011). Experiments showed that this conceptually simple method achieved state-of-the-art fair prediction performance and interpretability, showing its great potential for clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge