Genke Yang
TC-MGC: Text-Conditioned Multi-Grained Contrastive Learning for Text-Video Retrieval
Apr 07, 2025Abstract:Motivated by the success of coarse-grained or fine-grained contrast in text-video retrieval, there emerge multi-grained contrastive learning methods which focus on the integration of contrasts with different granularity. However, due to the wider semantic range of videos, the text-agnostic video representations might encode misleading information not described in texts, thus impeding the model from capturing precise cross-modal semantic correspondence. To this end, we propose a Text-Conditioned Multi-Grained Contrast framework, dubbed TC-MGC. Specifically, our model employs a language-video attention block to generate aggregated frame and video representations conditioned on the word's and text's attention weights over frames. To filter unnecessary similarity interactions and decrease trainable parameters in the Interactive Similarity Aggregation (ISA) module, we design a Similarity Reorganization (SR) module to identify attentive similarities and reorganize cross-modal similarity vectors and matrices. Next, we argue that the imbalance problem among multigrained similarities may result in over- and under-representation issues. We thereby introduce an auxiliary Similarity Decorrelation Regularization (SDR) loss to facilitate cooperative relationship utilization by similarity variance minimization on matching text-video pairs. Finally, we present a Linear Softmax Aggregation (LSA) module to explicitly encourage the interactions between multiple similarities and promote the usage of multi-grained information. Empirically, TC-MGC achieves competitive results on multiple text-video retrieval benchmarks, outperforming X-CLIP model by +2.8% (+1.3%), +2.2% (+1.0%), +1.5% (+0.9%) relative (absolute) improvements in text-to-video retrieval R@1 on MSR-VTT, DiDeMo and VATEX, respectively. Our code is publicly available at https://github.com/JingXiaolun/TC-MGC.
An Empirical Study of Excitation and Aggregation Design Adaptions in CLIP4Clip for Video-Text Retrieval
May 25, 2024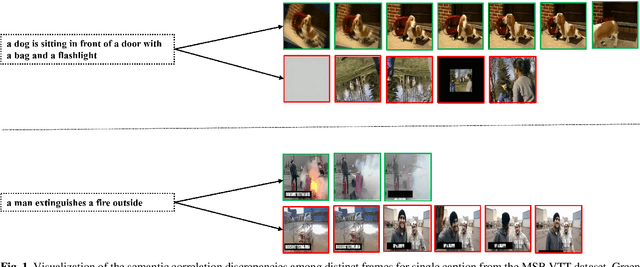

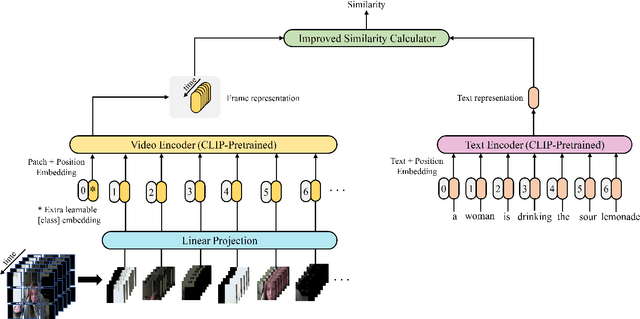
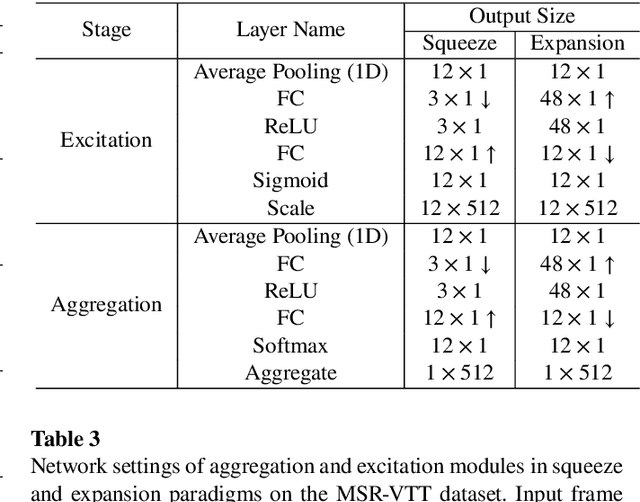
Abstract:CLIP4Clip model transferred from the CLIP has been the de-factor standard to solve the video clip retrieval task from frame-level input, triggering the surge of CLIP4Clip-based models in the video-text retrieval domain. In this work, we rethink the inherent limitation of widely-used mean pooling operation in the frame features aggregation and investigate the adaptions of excitation and aggregation design for discriminative video representation generation. We present a novel excitationand-aggregation design, including (1) The excitation module is available for capturing non-mutuallyexclusive relationships among frame features and achieving frame-wise features recalibration, and (2) The aggregation module is applied to learn exclusiveness used for frame representations aggregation. Similarly, we employ the cascade of sequential module and aggregation design to generate discriminative video representation in the sequential type. Besides, we adopt the excitation design in the tight type to obtain representative frame features for multi-modal interaction. The proposed modules are evaluated on three benchmark datasets of MSR-VTT, ActivityNet and DiDeMo, achieving MSR-VTT (43.9 R@1), ActivityNet (44.1 R@1) and DiDeMo (31.0 R@1). They outperform the CLIP4Clip results by +1.2% (+0.5%), +4.5% (+1.9%) and +9.5% (+2.7%) relative (absolute) improvements, demonstrating the superiority of our proposed excitation and aggregation designs. We hope our work will serve as an alternative for frame representations aggregation and facilitate future research.
The Graph Convolutional Network with Multi-representation Alignment for Drug Synergy Prediction
Nov 27, 2023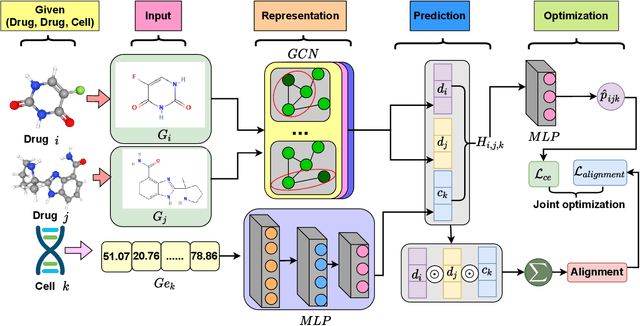
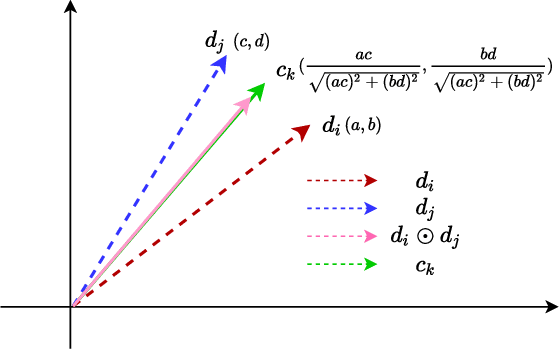
Abstract:Drug combination refers to the use of two or more drugs to treat a specific disease at the same time. It is currently the mainstream way to treat complex diseases. Compared with single drugs, drug combinations have better efficacy and can better inhibit toxicity and drug resistance. The computational model based on deep learning concatenates the representation of multiple drugs and the corresponding cell line feature as input, and the output is whether the drug combination can have an inhibitory effect on the cell line. However, this strategy of concatenating multiple representations has the following defects: the alignment of drug representation and cell line representation is ignored, resulting in the synergistic relationship not being reflected positionally in the embedding space. Moreover, the alignment measurement function in deep learning cannot be suitable for drug synergy prediction tasks due to differences in input types. Therefore, in this work, we propose a graph convolutional network with multi-representation alignment (GCNMRA) for predicting drug synergy. In the GCNMRA model, we designed a multi-representation alignment function suitable for the drug synergy prediction task so that the positional relationship between drug representations and cell line representation is reflected in the embedding space. In addition, the vector modulus of drug representations and cell line representation is considered to improve the accuracy of calculation results and accelerate model convergence. Finally, many relevant experiments were run on multiple drug synergy datasets to verify the effectiveness of the above innovative elements and the excellence of the GCNMRA model.
GraphCL-DTA: a graph contrastive learning with molecular semantics for drug-target binding affinity prediction
Jul 18, 2023



Abstract:Drug-target binding affinity prediction plays an important role in the early stages of drug discovery, which can infer the strength of interactions between new drugs and new targets. However, the performance of previous computational models is limited by the following drawbacks. The learning of drug representation relies only on supervised data, without taking into account the information contained in the molecular graph itself. Moreover, most previous studies tended to design complicated representation learning module, while uniformity, which is used to measure representation quality, is ignored. In this study, we propose GraphCL-DTA, a graph contrastive learning with molecular semantics for drug-target binding affinity prediction. In GraphCL-DTA, we design a graph contrastive learning framework for molecular graphs to learn drug representations, so that the semantics of molecular graphs are preserved. Through this graph contrastive framework, a more essential and effective drug representation can be learned without additional supervised data. Next, we design a new loss function that can be directly used to smoothly adjust the uniformity of drug and target representations. By directly optimizing the uniformity of representations, the representation quality of drugs and targets can be improved. The effectiveness of the above innovative elements is verified on two real datasets, KIBA and Davis. The excellent performance of GraphCL-DTA on the above datasets suggests its superiority to the state-of-the-art model.
Self-supervised Learning for Label Sparsity in Computational Drug Repositioning
Jun 01, 2022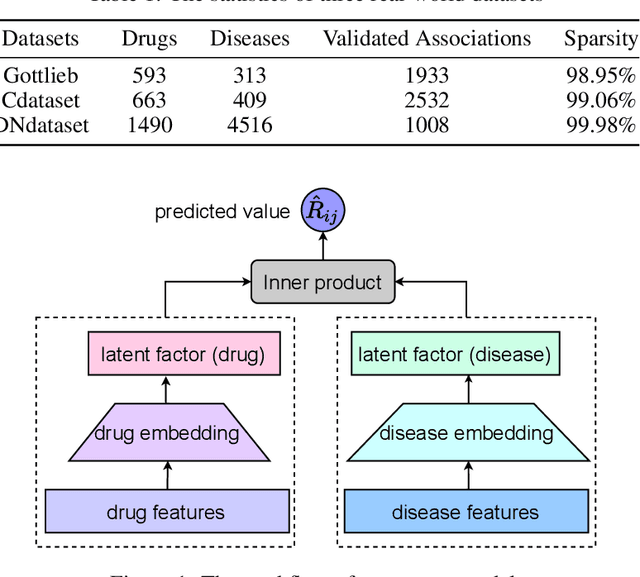
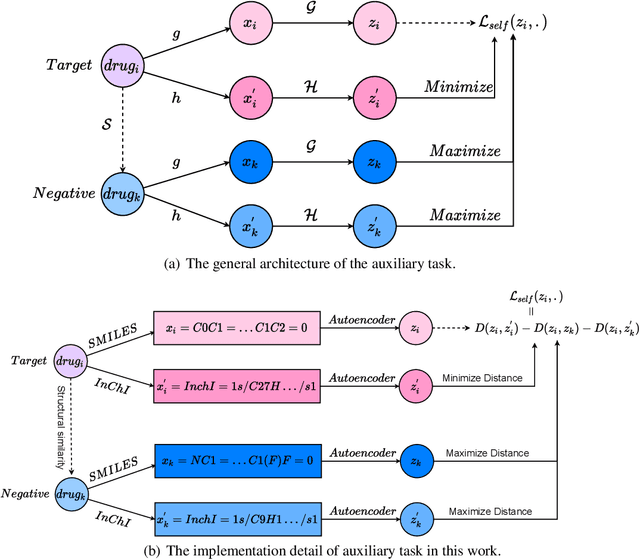
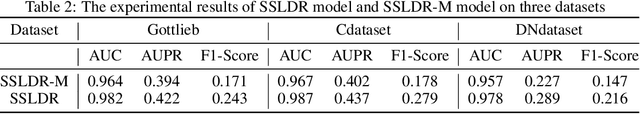
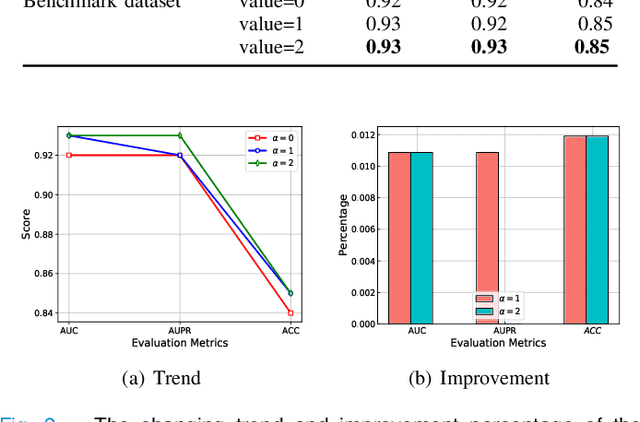
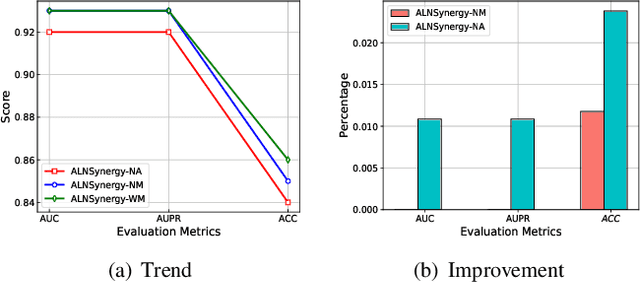
Abstract:The computational drug repositioning aims to discover new uses for marketed drugs, which can accelerate the drug development process and play an important role in the existing drug discovery system. However, the number of validated drug-disease associations is scarce compared to the number of drugs and diseases in the real world. Too few labeled samples will make the classification model unable to learn effective latent factors of drugs, resulting in poor generalization performance. In this work, we propose a multi-task self-supervised learning framework for computational drug repositioning. The framework tackles label sparsity by learning a better drug representation. Specifically, we take the drug-disease association prediction problem as the main task, and the auxiliary task is to use data augmentation strategies and contrast learning to mine the internal relationships of the original drug features, so as to automatically learn a better drug representation without supervised labels. And through joint training, it is ensured that the auxiliary task can improve the prediction accuracy of the main task. More precisely, the auxiliary task improves drug representation and serving as additional regularization to improve generalization. Furthermore, we design a multi-input decoding network to improve the reconstruction ability of the autoencoder model. We evaluate our model using three real-world datasets. The experimental results demonstrate the effectiveness of the multi-task self-supervised learning framework, and its predictive ability is superior to the state-of-the-art model.
The Computational Drug Repositioning without Negative Sampling
Nov 29, 2021
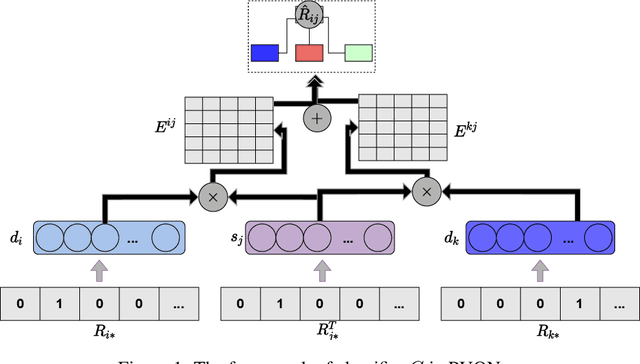
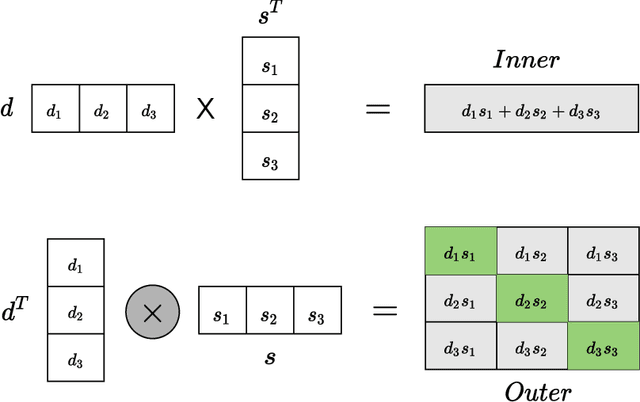
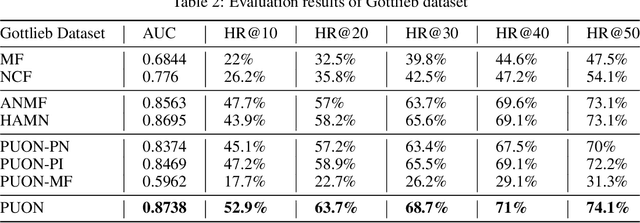
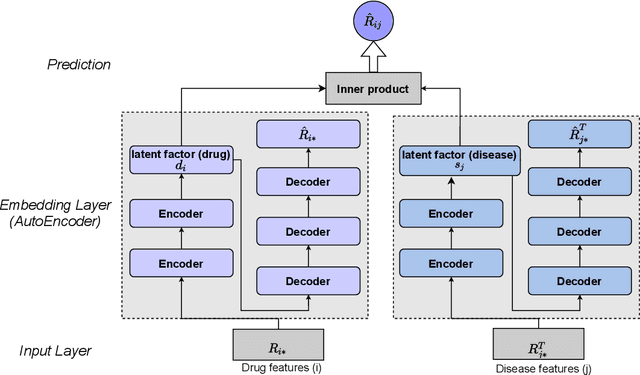
Abstract:Computational drug repositioning technology is an effective tool to accelerate drug development. Although this technique has been widely used and successful in recent decades, many existing models still suffer from multiple drawbacks such as the massive number of unvalidated drug-disease associations and inner product in the matrix factorization model. The limitations of these works are mainly due to the following two reasons: first, previous works used negative sampling techniques to treat unvalidated drug-disease associations as negative samples, which is invalid in real-world settings; Second, the inner product lacks modeling on the crossover information between dimensions of the latent factor. In this paper, we propose a novel PUON framework for addressing the above deficiencies, which models the joint distribution of drug-disease associations using validated and unvalidated drug-disease associations without employing negative sampling techniques. The PUON also modeled the cross-information of the latent factor of drugs and diseases using the outer product operation. For a comprehensive comparison, we considered 7 popular baselines. Extensive experiments in two real-world datasets showed that PUON achieved the best performance based on 6 popular evaluation metrics.
CenterFace: Joint Face Detection and Alignment Using Face as Point
Nov 09, 2019



Abstract:Face detection and alignment in unconstrained environment is always deployed on edge devices which have limited memory storage and low computing power. This paper proposes a one-stage method named CenterFace to simultaneously predict facial box and landmark location with real-time speed and high accuracy. The proposed method also belongs to the anchor free category. This is achieved by: (a) learning face existing possibility by the semantic maps, (b) learning bounding box, offsets and five landmarks for each position that potentially contains a face. Specifically, the method can run in real-time on a single CPU core and 200 FPS using NVIDIA 2080TI for VGA-resolution images, and can simultaneously achieve superior accuracy (WIDER FACE Val/Test-Easy: 0.935/0.932, Medium: 0.924/0.921, Hard: 0.875/0.873 and FDDB discontinuous: 0.980, continuous: 0.732). A demo of CenterFace can be available at https://github.com/Star-Clouds/CenterFace.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge