The Computational Drug Repositioning without Negative Sampling
Paper and Code
Nov 29, 2021
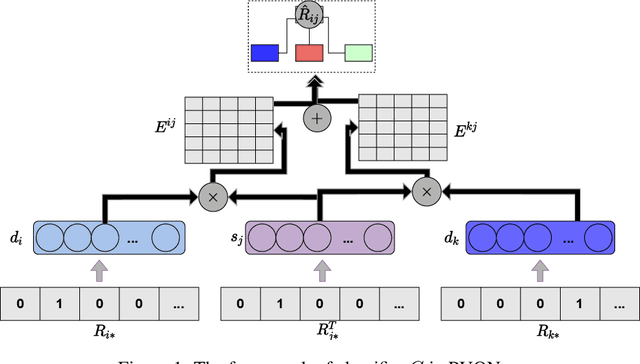
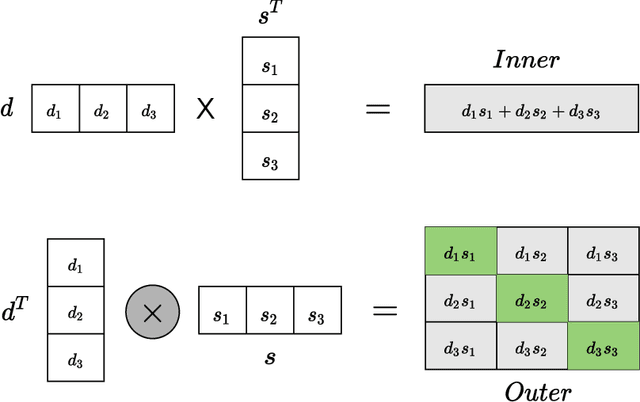
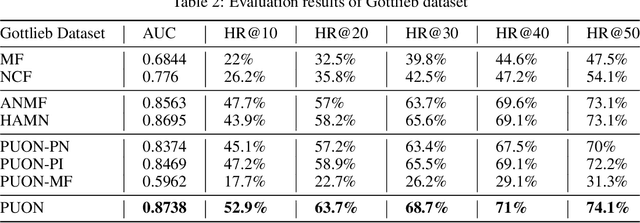
Computational drug repositioning technology is an effective tool to accelerate drug development. Although this technique has been widely used and successful in recent decades, many existing models still suffer from multiple drawbacks such as the massive number of unvalidated drug-disease associations and inner product in the matrix factorization model. The limitations of these works are mainly due to the following two reasons: first, previous works used negative sampling techniques to treat unvalidated drug-disease associations as negative samples, which is invalid in real-world settings; Second, the inner product lacks modeling on the crossover information between dimensions of the latent factor. In this paper, we propose a novel PUON framework for addressing the above deficiencies, which models the joint distribution of drug-disease associations using validated and unvalidated drug-disease associations without employing negative sampling techniques. The PUON also modeled the cross-information of the latent factor of drugs and diseases using the outer product operation. For a comprehensive comparison, we considered 7 popular baselines. Extensive experiments in two real-world datasets showed that PUON achieved the best performance based on 6 popular evaluation metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge