Self-supervised Learning for Label Sparsity in Computational Drug Repositioning
Paper and Code
Jun 01, 2022
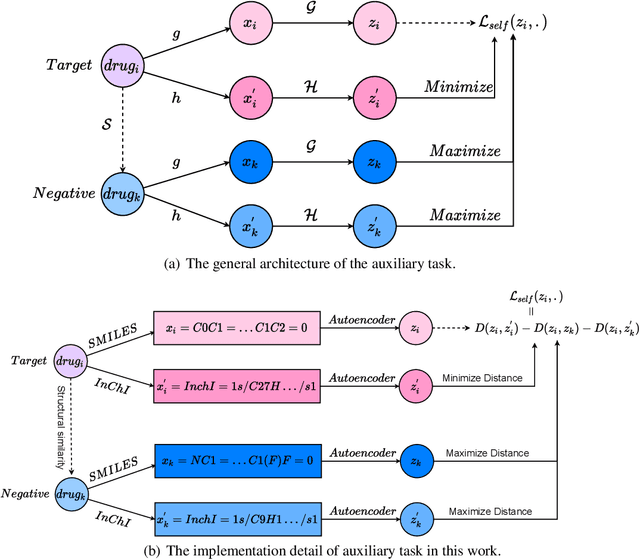
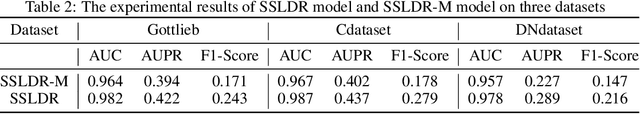
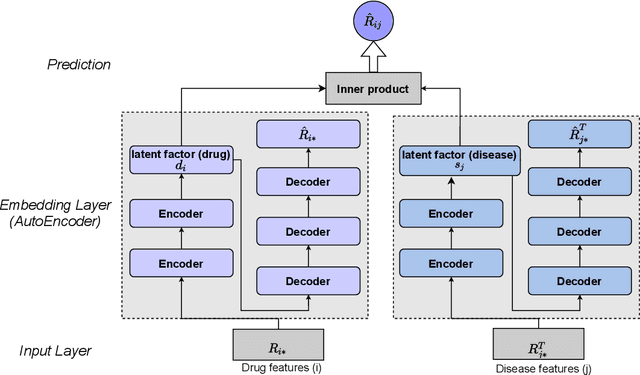
The computational drug repositioning aims to discover new uses for marketed drugs, which can accelerate the drug development process and play an important role in the existing drug discovery system. However, the number of validated drug-disease associations is scarce compared to the number of drugs and diseases in the real world. Too few labeled samples will make the classification model unable to learn effective latent factors of drugs, resulting in poor generalization performance. In this work, we propose a multi-task self-supervised learning framework for computational drug repositioning. The framework tackles label sparsity by learning a better drug representation. Specifically, we take the drug-disease association prediction problem as the main task, and the auxiliary task is to use data augmentation strategies and contrast learning to mine the internal relationships of the original drug features, so as to automatically learn a better drug representation without supervised labels. And through joint training, it is ensured that the auxiliary task can improve the prediction accuracy of the main task. More precisely, the auxiliary task improves drug representation and serving as additional regularization to improve generalization. Furthermore, we design a multi-input decoding network to improve the reconstruction ability of the autoencoder model. We evaluate our model using three real-world datasets. The experimental results demonstrate the effectiveness of the multi-task self-supervised learning framework, and its predictive ability is superior to the state-of-the-art model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge