Gary F. Egan
Towards Lower-Dose PET using Physics-Based Uncertainty-Aware Multimodal Learning with Robustness to Out-of-Distribution Data
Jul 21, 2021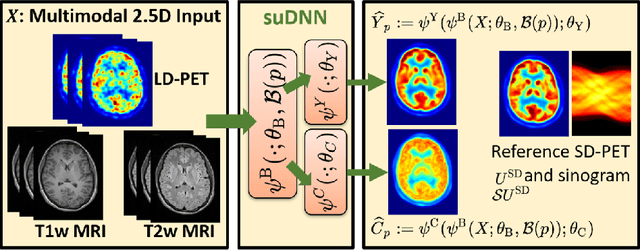
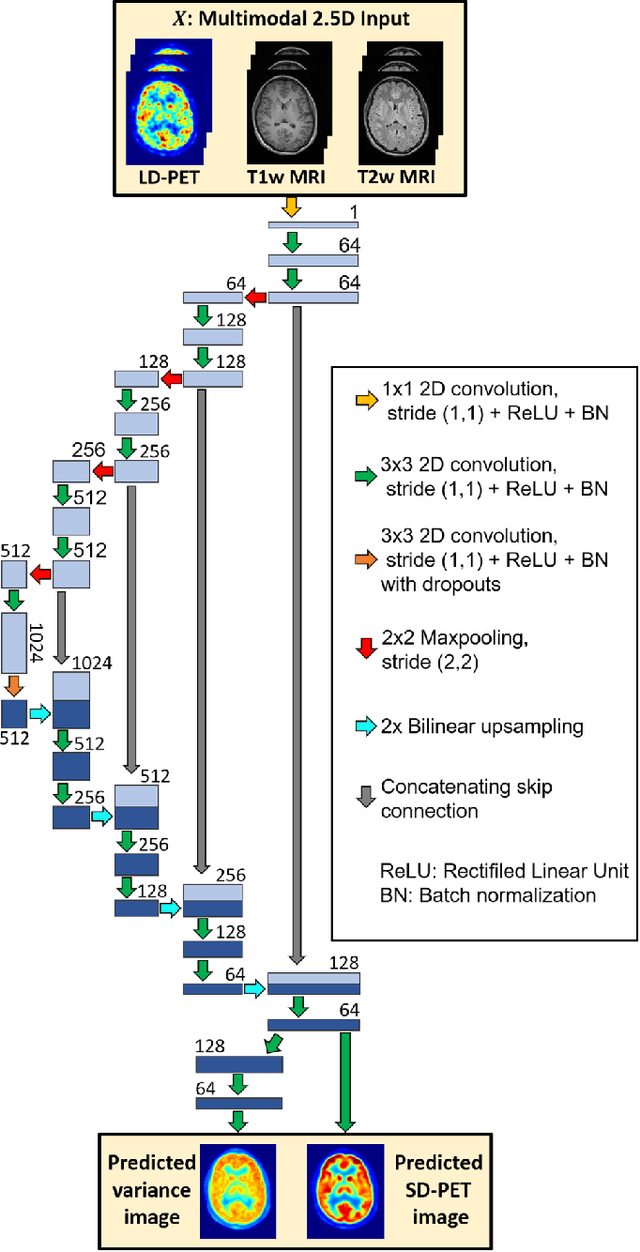
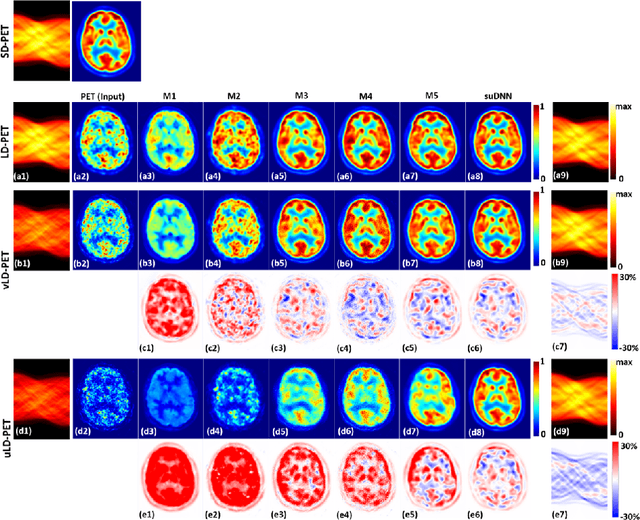
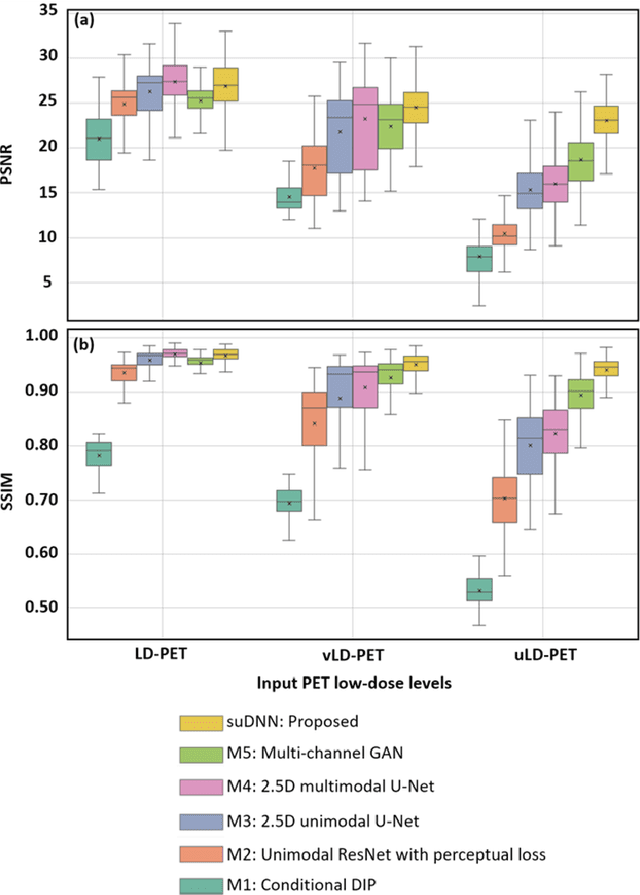
Abstract:Radiation exposure in positron emission tomography (PET) imaging limits its usage in the studies of radiation-sensitive populations, e.g., pregnant women, children, and adults that require longitudinal imaging. Reducing the PET radiotracer dose or acquisition time reduces photon counts, which can deteriorate image quality. Recent deep-neural-network (DNN) based methods for image-to-image translation enable the mapping of low-quality PET images (acquired using substantially reduced dose), coupled with the associated magnetic resonance imaging (MRI) images, to high-quality PET images. However, such DNN methods focus on applications involving test data that match the statistical characteristics of the training data very closely and give little attention to evaluating the performance of these DNNs on new out-of-distribution (OOD) acquisitions. We propose a novel DNN formulation that models the (i) underlying sinogram-based physics of the PET imaging system and (ii) the uncertainty in the DNN output through the per-voxel heteroscedasticity of the residuals between the predicted and the high-quality reference images. Our sinogram-based uncertainty-aware DNN framework, namely, suDNN, estimates a standard-dose PET image using multimodal input in the form of (i) a low-dose/low-count PET image and (ii) the corresponding multi-contrast MRI images, leading to improved robustness of suDNN to OOD acquisitions. Results on in vivo simultaneous PET-MRI, and various forms of OOD data in PET-MRI, show the benefits of suDNN over the current state of the art, quantitatively and qualitatively.
MoCoNet: Motion Correction in 3D MPRAGE images using a Convolutional Neural Network approach
Jul 29, 2018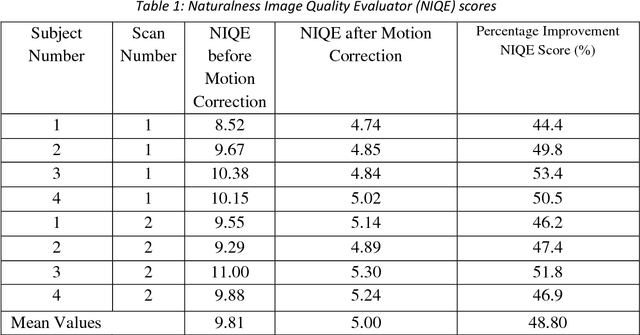
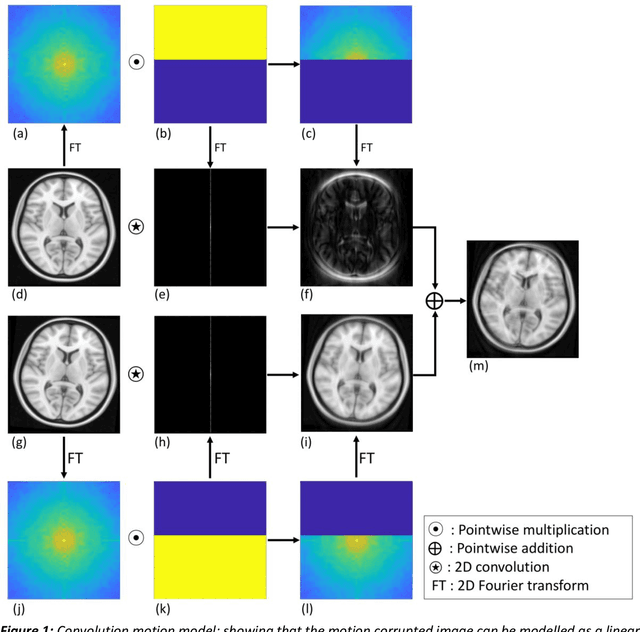
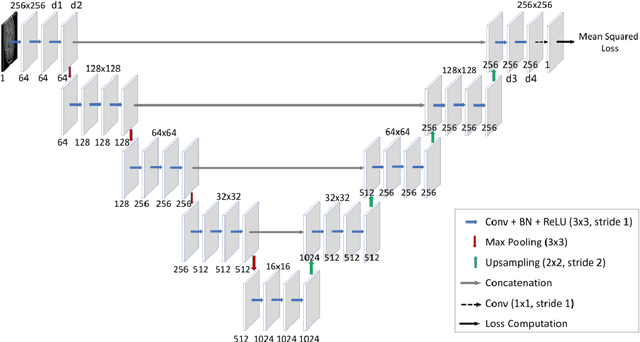
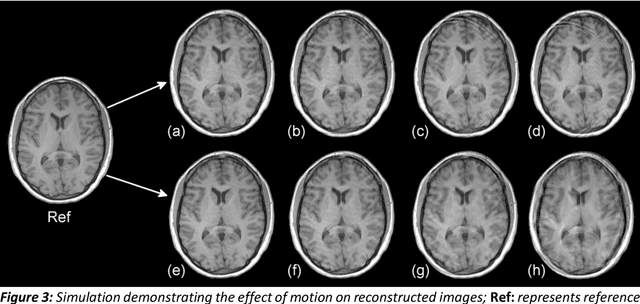
Abstract:Purpose: The suppression of motion artefacts from MR images is a challenging task. The purpose of this paper is to develop a standalone novel technique to suppress motion artefacts from MR images using a data-driven deep learning approach. Methods: A deep learning convolutional neural network (CNN) was developed to remove motion artefacts in brain MR images. A CNN was trained on simulated motion corrupted images to identify and suppress artefacts due to the motion. The network was an encoder-decoder CNN architecture where the encoder decomposed the motion corrupted images into a set of feature maps. The feature maps were then combined by the decoder network to generate a motion-corrected image. The network was tested on an unseen simulated dataset and an experimental, motion corrupted in vivo brain dataset. Results: The trained network was able to suppress the motion artefacts in the simulated motion corrupted images, and the mean percentage error in the motion corrected images was 2.69 % with a standard deviation of 0.95 %. The network was able to effectively suppress the motion artefacts from the experimental dataset, demonstrating the generalisation capability of the trained network. Conclusion: A novel and generic motion correction technique has been developed that can suppress motion artefacts from motion corrupted MR images. The proposed technique is a standalone post-processing method that does not interfere with data acquisition or reconstruction parameters, thus making it suitable for a multitude of MR sequences.
Multichannel Compressive Sensing MRI Using Noiselet Encoding
Jul 22, 2014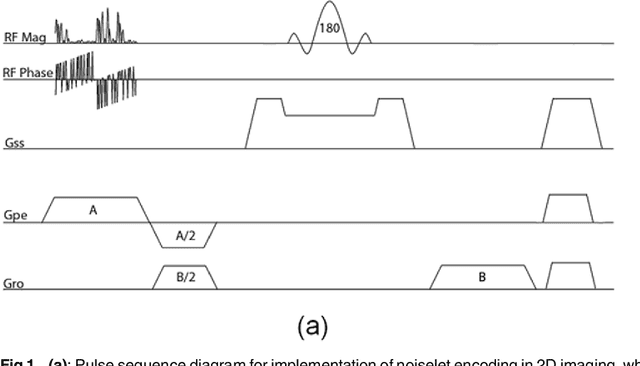
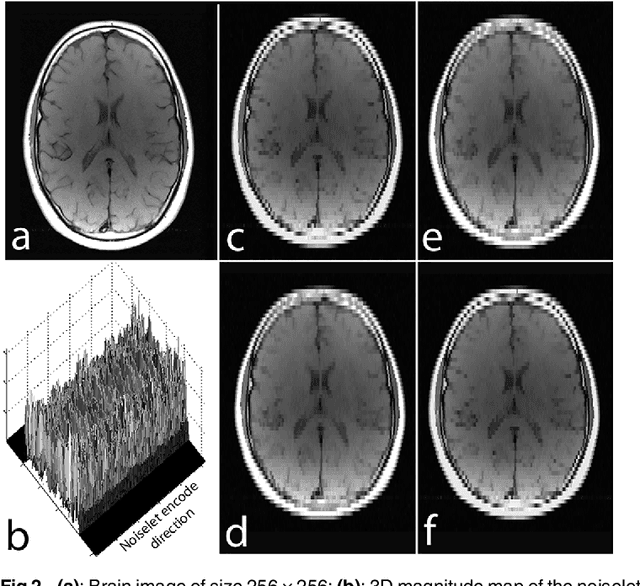
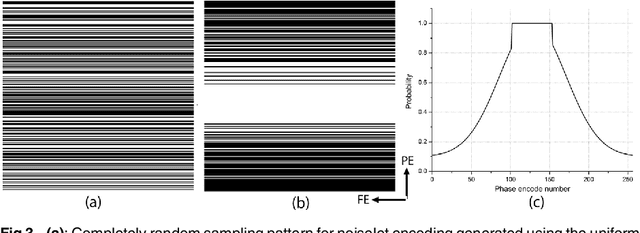
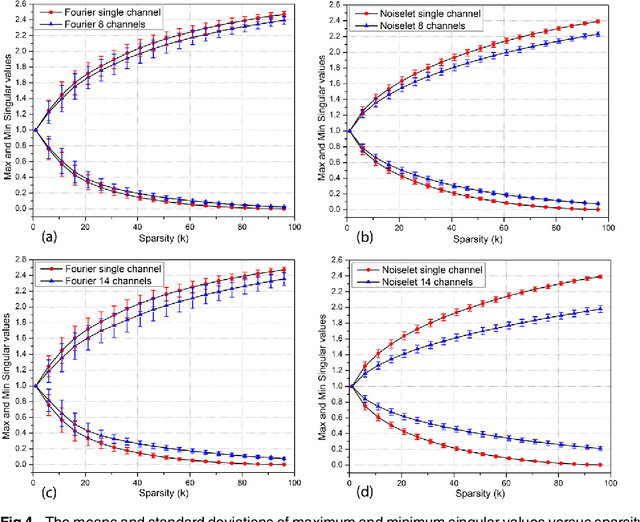
Abstract:The incoherence between measurement and sparsifying transform matrices and the restricted isometry property (RIP) of measurement matrix are two of the key factors in determining the performance of compressive sensing (CS). In CS-MRI, the randomly under-sampled Fourier matrix is used as the measurement matrix and the wavelet transform is usually used as sparsifying transform matrix. However, the incoherence between the randomly under-sampled Fourier matrix and the wavelet matrix is not optimal, which can deteriorate the performance of CS-MRI. Using the mathematical result that noiselets are maximally incoherent with wavelets, this paper introduces the noiselet unitary bases as the measurement matrix to improve the incoherence and RIP in CS-MRI, and presents a method to design the pulse sequence for the noiselet encoding. This novel encoding scheme is combined with the multichannel compressive sensing (MCS) framework to take the advantage of multichannel data acquisition used in MRI scanners. An empirical RIP analysis is presented to compare the multichannel noiselet and multichannel Fourier measurement matrices in MCS. Simulations are presented in the MCS framework to compare the performance of noiselet encoding reconstructions and Fourier encoding reconstructions at different acceleration factors. The comparisons indicate that multichannel noiselet measurement matrix has better RIP than that of its Fourier counterpart, and that noiselet encoded MCS-MRI outperforms Fourier encoded MCS-MRI in preserving image resolution and can achieve higher acceleration factors. To demonstrate the feasibility of the proposed noiselet encoding scheme, two pulse sequences with tailored spatially selective RF excitation pulses was designed and implemented on a 3T scanner to acquire the data in the noiselet domain from a phantom and a human brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge