Francesco Prinzi
Rad4XCNN: a new agnostic method for post-hoc global explanation of CNN-derived features by means of radiomics
Apr 26, 2024



Abstract:In the last years, artificial intelligence (AI) in clinical decision support systems (CDSS) played a key role in harnessing machine learning and deep learning architectures. Despite their promising capabilities, the lack of transparency and explainability of AI models poses significant challenges, particularly in medical contexts where reliability is a mandatory aspect. Achieving transparency without compromising predictive accuracy remains a key challenge. This paper presents a novel method, namely Rad4XCNN, to enhance the predictive power of CNN-derived features with the interpretability inherent in radiomic features. Rad4XCNN diverges from conventional methods based on saliency map, by associating intelligible meaning to CNN-derived features by means of Radiomics, offering new perspectives on explanation methods beyond visualization maps. Using a breast cancer classification task as a case study, we evaluated Rad4XCNN on ultrasound imaging datasets, including an online dataset and two in-house datasets for internal and external validation. Some key results are: i) CNN-derived features guarantee more robust accuracy when compared against ViT-derived and radiomic features; ii) conventional visualization map methods for explanation present several pitfalls; iii) Rad4XCNN does not sacrifice model accuracy for their explainability; iv) Rad4XCNN provides global explanation insights enabling the physician to analyze the model outputs and findings. In addition, we highlight the importance of integrating interpretability into AI models for enhanced trust and adoption in clinical practice, emphasizing how our method can mitigate some concerns related to explainable AI methods.
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023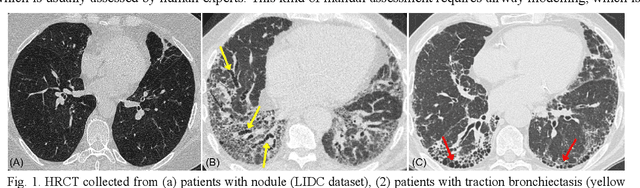
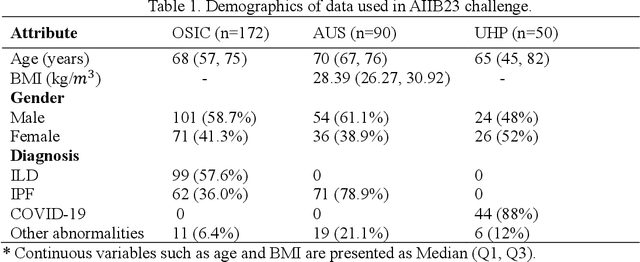
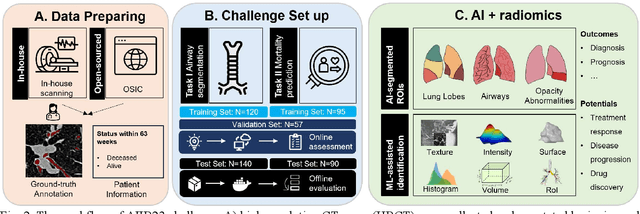
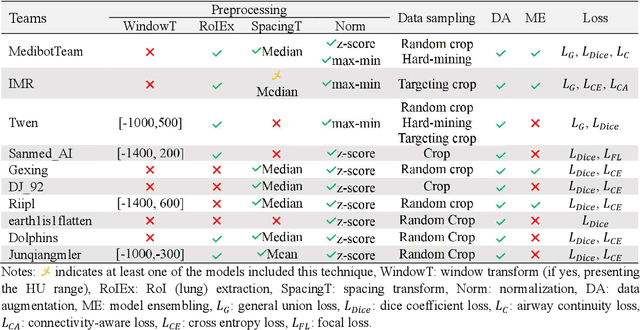
Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge