Felix Hohne
Large Scale Learning on Non-Homophilous Graphs: New Benchmarks and Strong Simple Methods
Oct 27, 2021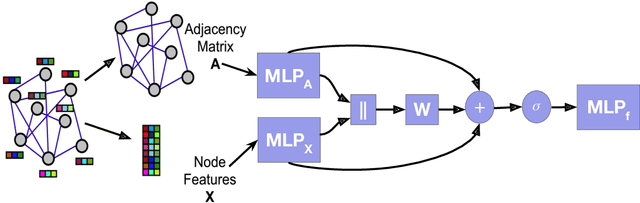
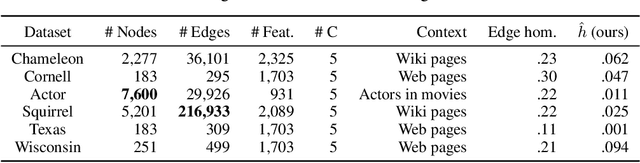
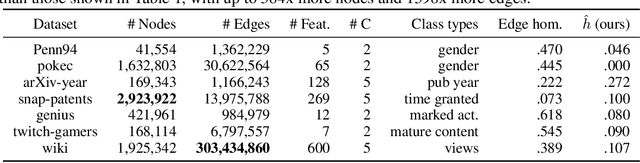
Abstract:Many widely used datasets for graph machine learning tasks have generally been homophilous, where nodes with similar labels connect to each other. Recently, new Graph Neural Networks (GNNs) have been developed that move beyond the homophily regime; however, their evaluation has often been conducted on small graphs with limited application domains. We collect and introduce diverse non-homophilous datasets from a variety of application areas that have up to 384x more nodes and 1398x more edges than prior datasets. We further show that existing scalable graph learning and graph minibatching techniques lead to performance degradation on these non-homophilous datasets, thus highlighting the need for further work on scalable non-homophilous methods. To address these concerns, we introduce LINKX -- a strong simple method that admits straightforward minibatch training and inference. Extensive experimental results with representative simple methods and GNNs across our proposed datasets show that LINKX achieves state-of-the-art performance for learning on non-homophilous graphs. Our codes and data are available at https://github.com/CUAI/Non-Homophily-Large-Scale.
New Benchmarks for Learning on Non-Homophilous Graphs
Apr 03, 2021



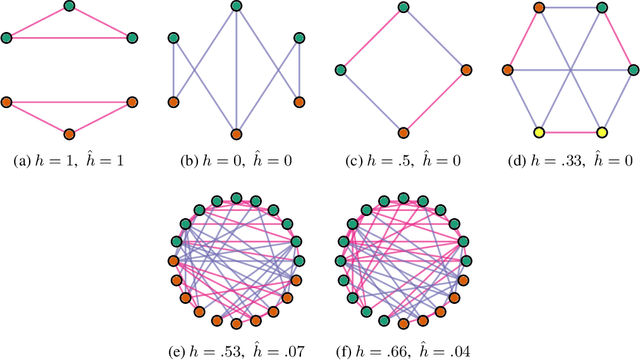
Abstract:Much data with graph structures satisfy the principle of homophily, meaning that connected nodes tend to be similar with respect to a specific attribute. As such, ubiquitous datasets for graph machine learning tasks have generally been highly homophilous, rewarding methods that leverage homophily as an inductive bias. Recent work has pointed out this particular focus, as new non-homophilous datasets have been introduced and graph representation learning models better suited for low-homophily settings have been developed. However, these datasets are small and poorly suited to truly testing the effectiveness of new methods in non-homophilous settings. We present a series of improved graph datasets with node label relationships that do not satisfy the homophily principle. Along with this, we introduce a new measure of the presence or absence of homophily that is better suited than existing measures in different regimes. We benchmark a range of simple methods and graph neural networks across our proposed datasets, drawing new insights for further research. Data and codes can be found at https://github.com/CUAI/Non-Homophily-Benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge