Felix Fuentes-Hurtado
Generalizable Denoising of Microscopy Images using Generative Adversarial Networks and Contrastive Learning
Mar 29, 2023Abstract:Microscopy images often suffer from high levels of noise, which can hinder further analysis and interpretation. Content-aware image restoration (CARE) methods have been proposed to address this issue, but they often require large amounts of training data and suffer from over-fitting. To overcome these challenges, we propose a novel framework for few-shot microscopy image denoising. Our approach combines a generative adversarial network (GAN) trained via contrastive learning (CL) with two structure preserving loss terms (Structural Similarity Index and Total Variation loss) to further improve the quality of the denoised images using little data. We demonstrate the effectiveness of our method on three well-known microscopy imaging datasets, and show that we can drastically reduce the amount of training data while retaining the quality of the denoising, thus alleviating the burden of acquiring paired data and enabling few-shot learning. The proposed framework can be easily extended to other image restoration tasks and has the potential to significantly advance the field of microscopy image analysis.
CaDIS: Cataract Dataset for Image Segmentation
Jul 19, 2019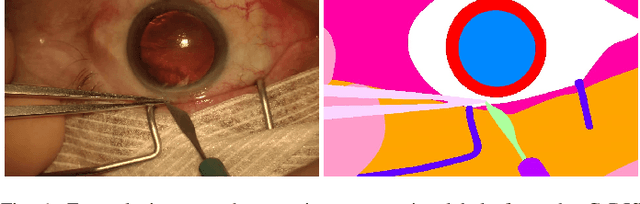
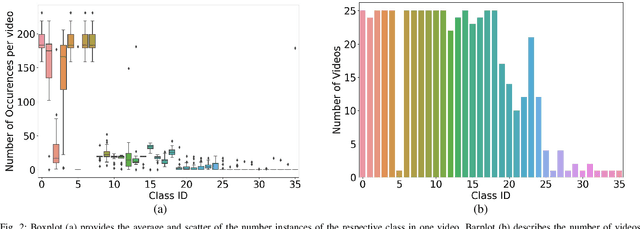
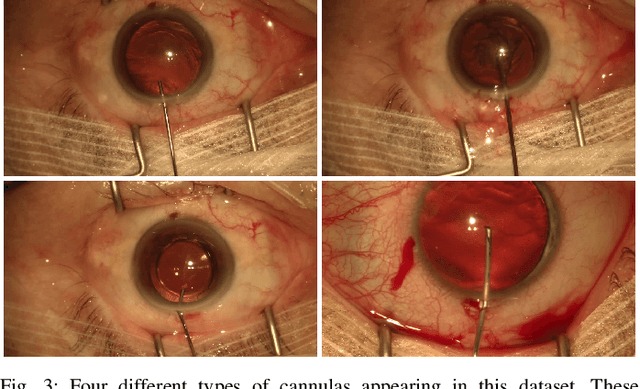
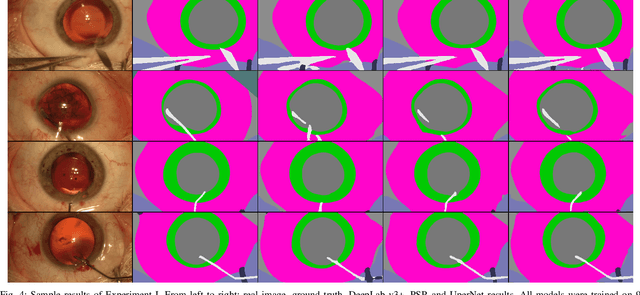
Abstract:Video signals provide a wealth of information about surgical procedures and are the main sensory cue for surgeons. Video processing and understanding can be used to empower computer assisted interventions (CAI) as well as the development of detailed post-operative analysis of the surgical intervention. A fundamental building block to such capabilities is the ability to understand and segment video into semantic labels that differentiate and localize tissue types and different instruments. Deep learning has advanced semantic segmentation techniques dramatically in recent years but is fundamentally reliant on the availability of labelled datasets used to train models. In this paper, we introduce a high quality dataset for semantic segmentation in Cataract surgery. We generated this dataset from the CATARACTS challenge dataset, which is publicly available. To the best of our knowledge, this dataset has the highest quality annotation in surgical data to date. We introduce the dataset and then show the automatic segmentation performance of state-of-the-art models on that dataset as a benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge